MAJOR UPDATE
As of Fall 2021, this wiki has been discontinued and is no longer being actively developed.
All updated materials and announcements for the QCBS R Workshop Series are now housed on the QCBS R Workshop website. Please update your bookmarks accordingly to avoid outdated material and/or broken links.
Thank you for your understanding,
Your QCBS R Workshop Coordinators.
QCBS R Workshops
This series of 10 workshops walks participants through the steps required to use R for a wide array of statistical analyses relevant to research in biology and ecology. These open-access workshops were created by members of the QCBS both for members of the QCBS and the larger community.
The content of this workshop has been peer-reviewed by several QCBS members. If you would like to suggest modifications, please contact the current series coordinators, listed on the main wiki page
Workshop 1: Introduction to R
Developed by: Sylvain Christin, Cédric Frenette Dussault, Dalal Hanna
Summary: In this introductory R Workshop you will learn what R open-source statistical software is, why you should absolutely start using it, and all the first steps to help you get started in R. We will show you how R can act as a calculator, teach you about the various types of objects in R, show you how to use functions and load packages, and find all the resources you need to get help. If any of this sounds obscure, don’t worry! By the end of this workshop you’ll know what all these words mean!
Link to new Rmarkdown presentation
Please try it and tell the R workshop coordinators what you think!
Link to old Prezi presentation
Download the R script for this lesson.
Installing R and RStudio
Before you can effectively start this workshop, you will need to install the R and RStudio softwares on your computer. For R, go to http://www.r-project.org/ and click on download. You will have to select a mirror site (usually one close to you) and select your platform (OS X, Windows or Linux). Download the file and launch it to complete the installation. You don't have to change the default settings.
To install RStudio, go to http://www.rstudio.com/ and select RStudio from the Products tab. Click on the open source version of RStudio Desktop and select your platform to download it. Launch the file you just downloaded to complete the installation. Again, you can keep the default settings.
Learning Objectives
- Understand what R and RStudio are;
- Use R as a calculator;
- Manipulate objects in R;
- Install, use an R package and get help.
Understand what R and RStudio are
What is R?
- R is an open source programming language and environment.
- It is designed for data analysis, graphical display and data simulations.
- It is one of the world's leading statistical programming environment.
Why use R?
- R is open-source! This means that it is free, and constantly being updated and improved.
- It is compatible. R works on most existing operating systems.
- R can help you create tables, produce graphs and do your statistics, all within the same program. So with R, there is no need to use more than one program to manage data for your publications. Everything can happen in one single program.
- More and more scientists are using R every year. This means that its capacities are constantly growing and will continue to increase over time. This also means that there is a big online community of people that can help with any problems you have in R.
Using RStudio
RStudio is an integrated development environment (IDE) for R. Basically, it's a place where you can easily use the R language, visualize tables and figures and even run all your statistical analyses. We recommend using it instead of the traditional command line as it provides great visual aid and a number of useful tools that you will learn more about over the course of this workshop.
CHALLENGE 1
Open RStudio
Note for Windows users: If the restriction “unable to write on disk” appears when you try to open R-Studio, right-click on your R-Studio icon and chose:“Execute as administrator” to open the program.
The console
When you open RStudio, the first thing that you see to the left of the screen is the “console”. This is where we will be working for the rest of this Introduction to R workshop.
Text in the console typically looks like this:
- Illustrating R console input and output
> output [1] "This is the output"
Note 1: You always have to push “enter” for the input to run in the console.
Note 2: People often wonder what the brackets in front of the output mean. They are there to help you locate “where” you are in the output. For example, if you ask R to output numbers between 1 and 10 and the output is on 2 rows, the bracket at the start of the 2nd row will help you understand at which value of the output you are situated:
- Understanding the brackets in front of the console output
[1] 1 2 3 4 5 [6] 6 7 8 9 10
2. Use R as a calculator
The first thing to know about the R console is that you can use it as a calculator.
- Addition
> 1 + 1 [1] 2
- Substraction
> 10 - 1 [1] 9
- Multiplication
> 2 * 2 [1] 4
- Division
> 8 / 2 [1] 4
- Exponents
> 2^3 [1] 8
CHALLENGE 2
Use R to calculate the following skill testing question: 2 + 16 x 24 - 56
CHALLENGE 3
Use R to calculate the following skill testing question:
2 + 16 x 24 - 56 / (2 + 1) - 457
Pay attention to the order of operations when thinking about this question!
Note that R always follows the order of priorities.
R TIP
Try using the “up” and “down” arrows to reproduce previous commands. These keys actually allow you to scroll through your command history. This is a useful tool to go back and see what command you ran and if you might have made a mistake in it. This is always a useful tool to quickly alter previous commands you ran and to re-run them in a slightly different way.
CHALLENGE 4
What is the area of a circle with a radius of 5 cm?
3. Manipulate objects in R
Object
You have learned so far how to use R as a calculator to obtain various numerical values. However, it can get tiresome to always write the same code down in the R console, especially if you have to use some values repeatedly. This is where the concept of object becomes useful.
R is an object-oriented programming language. What this means is that we can allocate a name to values we've created to save them in our workspace. An object is composed of three parts: 1) a value we're interested in, 2) an identifier and 3) the assignment operator.
- The value can be almost anything we want: a number, the result of a calculation, a string of characters, a data frame, a plot or a function.
- The identifier is the name you assign to the value. Whenever you want to refer to this value, you simply type the identifier in the R console and R will return its value. Identifiers can include only letters, numbers, periods and underscores, and should always begin with a letter.
- The assignment operator resembles an arrow (
<-) and is used to link the value to the identifier.
The following code clarifies these ideas:
- Illustrating the concept of object
# Let's create an object called mean_x. # The # symbol is used in R to indicate comments. It is not processed by R. # It is important to add comments to code so that it can be understood and used by other people. mean_x <- (2 + 6) / 2 # Typing its name will return its value. > mean_x [1] 4
Here, (2 + 6) / 2 is the value you want to save as an object. The identifier mean_x is assigned to this value. Typing mean_x returns the value of the calculation (i.e. 4). You have to be scrupulous when typing the identifier because R is case-sensitive: writing mean_x is not the same as writing MEAN_X. You can see that the assignment operator <- creates an explicit link between the value and the identifier. It always points from the value to the identifier. Note that it is also possible to use the equal sign = as the assignment operator but it is preferable not to because it is also used for other operations in R, which can cause problems when using it for assignment. Finally, imagine that the operator <- and = follow their own order of priorities.
- Order of priorities with assignment operator and equal sign
> y <- x = 5 Error in y <- x = 5 : object 'y' not found > y = x <- 5 > y [1] 5 > x [1] 5
Good practices in R code
Name
- Try having short and explicit names for your variables. Naming a variable
varis not very informative. - Use an underscore (
_), or a dot (.) to separate words within a name and try to be consistent! - Avoid using names of existing functions and variables (e.g.,
c,table,T, etc.)
Space
- Add spaces around all operators (
=,+,-,←, etc.) to make the code more readable. - Always put a space after a comma, and never before (like in regular English).
CHALLENGE 5
Create an object with a value of 1 + 1.718282 (Euler's number) and name iteuler_value
CHALLENGE 6
Create an object (you decide the value) with a name that starts with a number. What happens?
R TIP
Using the Tab key allows auto-completion of names. It speeds up command entering and avoids spelling errors. For example, if you type eu and then press tab, you will see a list of objects or functions beginning with eu. Select euler_value (the object we just created) and press enter. The euler_value identifier now appears at the command line.
Types of data structures in R
Using R to analyze your data is an important aspect of this software. Data comes in different forms and can be grouped in distinct categories. Depending on the nature of the values enclosed inside your data or object, R classifies them accordingly. The following figure illustrates common objects found in R.
The first object is a vector. It is one of the most common objects in R. A vector is an entity consisting of a list of related values. All values in a vector must be the same mode. The main modes in R are numeric, character and logical. Numeric vectors are made of numbers only. Character vectors include text strings or a mix of text strings and other modes. You need to use "" to delimit elements in a character vector. Logical vectors include TRUE/FALSE entries only. A vector with a single value (usually a constant) is called an atomic vector.
Before we look at how to create different types of vectors, let's have a look at the generic method of creating vectors. If you recall what you have just learned, you will first have to identify some value you want to put in a vector and then link it to an identifier with the assignment operator (i.e. create an object). When you have more than one value in a vector, you need a way to tell R to group all these values to create a vector. The trick here is to use the c() function. Don't worry, you will learn about functions pretty soon in one of the following sections. For now, just remember to put your values between parentheses next to letter c() in this format: vector.name <- c(value1, value2, value3, …). The function c() means combine or concatenate. It is a quick and easy function so remember it!
Now that you know the generic method to create a vector in R, let's have a look at how to create different types of vectors.
- Creating vectors in R
# Create a numeric vector with the c (which means combine or concatenate) function. # We will learn about functions soon! > num_vector <- c(1, 4, 3, 98, 32, -76, -4) # Create a character vector. Always use "" to delimit text strings! > char_vector <- c("blue", "red", "green") # Create a logical or boolean vector. Don't use "" or R will consider this as text strings. > bool_vector <- c(TRUE, TRUE, FALSE) #It is also possible to use abbreviations for logical vectors. > bool_vector2 <- c(T, T, F)
CHALLENGE 7
Create a vector containing the first five odd numbers (starting from 1) and name it odd_n.
R TIP
Use dput() function to obtain the reverse, i.e. the content of an object formatted as a vector. e.g. :
> odd_n <- c(1, 3, 5, 7, 9) > odd_n [1] 1 3 5 7 9 > dput(odd_n) c(1, 3, 5, 7, 9) # The output can be copied-pasted to create a new object by using the structure() function > structure(c(1, 3, 5, 7, 9))
This demonstration might not be that convincing, but keep in mind that it can be very useful when you're manipulating data. These functions are really useful to provide a reproducible example for a question on Stack Overflow for instance (see one more application in the part about data frames)!
What you have learned previously about calculations is also valid for vectors: vectors can be used for calculations. The only difference is that when a vector has more than 1 element, the operation is applied on all elements of the vector. The following example clarifies this.
- Calculations with vectors
# Create two numeric vectors. > x <- 1:5 # An equivalent form is: x <- c(1:5). > y <- 6 # Remember that the : symbol, when used with numbers, is the sequence operator. # It tells R to create a series of numbers increasing by 1. # Equivalent to this is x <- c(1,2,3,4,5) # Let's sum both vectors. # 6 is added to all elements of the x vector. > x + y [1] 7 8 9 10 11 # Let's multiply x by itself. > x * y [1] 6 12 18 24 30
Another important type of object you will use regularly is the data frame. A data frame is a group of vectors of the same length (i.e. the same number of elements). Columns are always variables and rows are observations, cases, sites or replicates. Different modes can be saved in different columns (but always the same mode in a column). It is in this format that ecological data are usually stored. The following example shows a fictitious dataset representing 4 sites where soil pH and the number of plant species were recorded. There is also a “Treatment” variable (fertilized or not). Let's have a look at the creation of a data frame.
| site_id | soil_pH | num_sp | treatment |
|---|---|---|---|
| A1.01 | 5.6 | 17 | Fertilised |
| A1.02 | 7.3 | 23 | Fertilised |
| B1.01 | 4.1 | 15 | Not Fertilised |
| B1.02 | 6.0 | 17 | Not Fertilised |
- Creating a data frame
# We first start by creating vectors. > site_id <- c("A1.01", "A1.02", "B1.01", "B1.02") > soil_pH <- c(5.6, 7.3, 4.1, 6.0) > num_sp <- c(17, 23, 15, 7) > treatment <- c("Fert", "Fert", "No_fert", "No_fert") # We then combine them to create a data frame with the data.frame function. my_df <- data.frame(site_id, soil_pH, num_sp, treatment) # Visualise it! my_df
R TIP
Here the dput() function in another example.
> dput(my_df) structure(list(site_id = c("A1.01", "A1.02", "B1.01", "B1.02" ), soil_pH = c(5.6, 7.3, 4.1, 6), num_sp = c(17, 23, 15, 7), treatment = c("Fert", "Fert", "No_fert", "No_fert")), class = "data.frame", row.names = c(NA, -4L)) # It's possible to rebuild the initial data frame (with some associated metadata as the class of variables) by copying and pasting the previous output: > structure(list(site_id = c("A1.01", "A1.02", "B1.01", "B1.02" ), soil_pH = c(5.6, 7.3, 4.1, 6), num_sp = c(17, 23, 15, 7), treatment = c("Fert", "Fert", "No_fert", "No_fert")), class = "data.frame", row.names = c(NA, -4L))
Other types of objects include matrices, arrays and lists. A matrix is similar to a data frame except that all cells in the matrix must be the same mode. An array is similar to a matrix but can have more than two dimensions. Arrays are usually used for advanced computation like numerical simulations and permutation tests. A list is an aggregation of various types of objects. For example, a list could include a vector, a data frame and a matrix in the same object.
Indexing objects in R
Indexing a vector
Typing an object's name in R returns the complete object. But what if our object is a huge data frame with millions of entries? It can easily become confusing to identify specific elements of an object. R allows us to extract only part of an object. This is called indexing. We specify the position of values we want to extract from an object with brackets [ ]. The following code illustrates the concept of indexation with vectors.
- Indexing a vector
# Let's first create a numeric and a character vector. # There is no need to do this again if you already did it in the previous exercise! > odd_n <- c(1, 3, 5, 7, 9) # Extract the second element of the numeric vector. > odd_n[2] [1] 3 # Extract the second and fourth elements of the numeric vector. > odd_n[c(2, 4)] [1] 3 7 # Extract all but the two first elements of the numeric vector. > odd_n[c(-1, -2)] [1] 5 7 9 # If you select a position that is not in the numeric vector > odd_n[c(1,6)] [1] 1 NA # There is no sixth value in this vector so R returns a null value (i.e. NA) # NA stands for 'Not available'. # You can use logical statement to select values. > odd_n[odd_n > 4] [1] 5 7 9 # Extract all elements of the character vector corresponding exactly to "blue". > char_vecteur[char_vecteur == "blue"] [1] "bleu" # Note the use of the double equal sign ==.
CHALLENGE 8
a) Extract the 4th value of thenum_vectorvector.
b) Extract the 1st and 3rd values of thenum_vectorvector.
c) Extract all values of thenum_vectorvector excluding the 2nd and 4th values.
Challenge 8a: Indexing vectors
Challenge 8b: Indexing vectors
Challenge 8c: Indexing vectors
CHALLENGE 9
Explore the difference between these 2 lines of code:
- Differences between codes
> char_vector == "blue" > char_vector[char.vector == "blue"]
Challenge 9: Differences between codes
Indexing a data frame
For data frames, the concept of indexation is similar, but we usually have to specify two dimensions: the row and column numbers. The R syntax is
dataframe[row number, column number].
Here are a few examples of data frame indexation. Note that the first four operations are also valid for indexing matrices.
- Indexing a data frame
# Let's reuse the data frame we created earlier (my_df) # Extract the 1st row of the data frame > my_df[1, ] # Extract the 3rd columm > my_df[, 3] # Extract the 2nd element of the 4th column > my_df[2, 4] # Extract lines 2 to 4 > my_df[c(2,4), ] # Extract the "site_id" column by referring directly to its name # The dollar sign ($) allows such an operation! > my_df$site_id # Extract the "site_id" and "soil_pH" variables > my_df[, c("site_id","soil_pH")]
A quick note on logical statements
R gives you the possibility to test logical statements, i.e. to evaluate whether a statement is true or false. You can compare objects with the following logical operators:
| Operator | Description |
|---|---|
| < | less than |
| <= | less than or equal to |
| > | greater than |
| >= | greater than or equal to |
| == | exactly equal to |
| != | not equal to |
| x | y | x OR y |
| x & y | x AND y |
The following examples illustrate how to use these operators properly.
- Testing logical statements
# First, let's create two vectors for comparison. > x2 <- c(1:5) > y2 <- c(1, 2, -7, 4, 5). # Let's verify if the elements in x2 are greater or equal to 3. # R returns a TRUE/FALSE value for each element (in order). > x2 >= 3 [1] FALSE FALSE TRUE TRUE TRUE # Let's see if the elements of x2 are exactly equal to those of y2. > x2 == y2 [1] TRUE TRUE FALSE TRUE TRUE # Is 3 not equal to 4? Of course! > 3 != 4 [1] TRUE # Let's see which values in x2 are greater than 2 but smaller than 5. # You have to write x2 twice. # If you write x2 > 2 & < 5, you will get an error message. > x2 > 2 & x2 < 5 [1] FALSE FALSE TRUE TRUE FALSE
CHALLENGE 10
a) Extract thenum_spcolumn frommy_dfand multiply its values by the first four values of thenum_vecvector.
b) After that, write a statement that checks if the values you obtained are greater than 25. Refer to challenge 9 to complete this challenge.
Challenge 10a: Indexing and multiplying
Challenge 10b: Logical statement
Functions
Most of the time with R, you will need to use functions to do what you want.
Functions are tools that are there to simplify your life. They allow you to quickly execute operations on objects without having to write every mathematical step. Functions are usually pre-existing R code that are executed when used. They remove the need to create this code and to rewrite it every time you need it.
To execute a function, you will need to call it. A function call is essentially a shortcut to the code of the function.
To perform the function call you will need entry values called arguments (or sometimes parameters). After performing its operations, the function will then give you a return value. The command also must be structured properly, following the “grammar rules” of the R language (syntax).
A function call is structured as follow: the name of the function, followed by parenthesis ( ). Inside the parenthesis, you insert all your arguments separated with commas.
function_name(arg1, arg2, …)
Here an example:
- Function syntax
> sum(1, 2)
Arguments are values and the instructions the function needs to run. Objects can be passed into functions:
- Objects as arguments
> a <- 3 > b <- 5 > sum(a, b) [1] 8
On the last line, the output that appears is the return value of the function. In this case, it is the sum of a and b, 8.
CHALLENGE 11
a) - Create a vector a that contains all numbers from 1 to 5
- Create an object b with value of 2
- Add a and b together using the basic+operator and save the result in an object calledresult_add
- Add a and b together using thesum()function and save the result in an object calledresult_sum
- Compare result_add and result_sum. Are they different?
b) Add 5 toresult_sumusing thesum()function.
Challenge 11a: Calling functions
Challenge 11b: Calling functions
Arguments
Arguments each have a name that can be provided during a function call.
If the name is not provided, the order of the arguments does matter.
If the name is provided, the order of the arguments does not matter.
To provide an argument name during a function call, just enter it in the form name=value.
- Argument name
log(8, base = 2)
—–
CHALLENGE 12
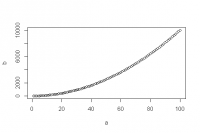
plot(x, y)is a function that draws a graph of y as a function of x. It requires two arguments namedxandy. What are the differences between the following lines?
- Challenge 12
> a <- 1:100 > b <- a^2 > plot(a, b) > plot(b, a) > plot(x = a, y = b) > plot(y = b, x = a)
As a reference, here is a list of some of the most common R functions:
sqrt, log, exp, max, min, sum, mean, sd, var, summary, plot, par, paste, format, head, length, str, names, typeof, class, attributes, library, ls, rm, setwd, getwd, file.choose, c, seq, rep, tapply, lapply, aggregate, merge, cbind, rbind, unique, help (or ?), help.search (or ??), help.start
4. Install, use an R package and get help
Package
Packages are a grouping of functions and/or datasets that share a similar theme. Ex : statistics, spatial analysis, plotting…
Everyone can develop packages and make them available to other R users.
They are usually available through the Comprehensive R Archive Network (CRAN)http://cran.r-project.org/web/packages/
Currently, more than 16000 packages are publicly available.
To install packages on your computer, use the function install.packages()
> install.packages("ggplot2")
Installing a package is required only once (but updated from time to time) but it is not enough to use its functions. You need to load the package once per R session before using it with the library() function.
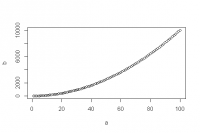
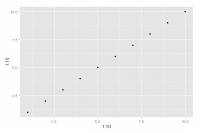
Let's try using the function qplot() found in the ggplot2 package we just installed.
> qplot(1:10, 1:10)
The package was correctly installed but we didn't load it. Therefore, the execution of this code leads to the following error:
Error: could not find function “qplot”
To be able to use the function qplot() we need to load the package ggplot2 before.
> library("ggplot2") > qplot(1:10, 1:10)
Now the function is found and the execution of our code leads to the following graph:
It is good practice to unload packages once we are done with them because it might conflict with other packages. Unloading a package is done with the detach() function and by specifying that it is a package:
- Unloading a package
> detach(package:ggplot2)
Getting help
Getting help with functions
We've seen so far that R is really great and offers us a lot of functions to work with. Among all these functions, there are probably some that can do what we want.
Now the problem is: how to find them?
To find a function that does something specific in your installed packages, you can use the operator ?? (or the help.search() function). To perform a search, just type ?? and what you want to search.
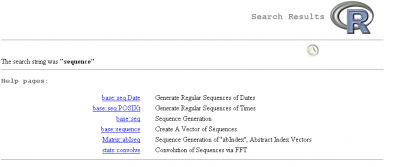
For example, let's say we want to create a sequence of odd numbers between 0 and 10. We will search in our packages all functions with the word sequence in them.
This opens the following window (Note: the search result may vary depending on the packages installed on your computer):
The search result contains two columns:
- On the left, we have the name of the function and the package in which we can find it in the format
package_name::function_name - On the right, we have the description of the function
Usually, functions have names that are representative of what they do. This makes it easier to use. Remember this if you ever start to write your own functions!
Here the result that interests us is base::seq, i.e. the function seq that can be found in the base package and that generates sequences.
Note: the base package contains basic functions that load with R when you launch it and are therefore always available.
Help page of functions
We will use the seq() function to generate our sequence. But how does it work? What does it do? How should I use it?
To answer all these questions, we'll try to find the help page of the function. For that, we will use the ? operator (or the help() function). To access the help page of a function, we enter the commage as follow:
?function_name
So for the seq() function, we type:
- Finding help
> ?seq
This opens the following page:
A help page usually contains the folowing elements and sections (Note: sometimes, the same help page is used for more than one function):
- On the top left corner, the name of the function and the package it belongs to in the format
function{package} - Description: a short description of the function(s)
- Usage: how to use the function(s), especially what are the name and order of the arguments. If a value is specified near an argument name, it means that a default value has been defined for this argument, which makes it optional to specify another value. If the argument is missing, the default value will be used. For example, if we do not provide a
fromargument, the sequence will automatically start from 1 - Arguments: A detailed description of all the arguments and what is expected or required for the function to work correctly. Be careful! Here are listed all the arguments for all the functions described on the help page; not all arguments are available for all listed functions. For example, in this help page, the arguments
fromandtoare not available for the functionseq_along(). - Details: Provide in depth details of the inner working of the function(s). Some specific cases can be discussed here or additionnal information provided.
- Value: Explains what the return value of the function is.
- References: Sources used as basis for the function or interesting readings on the subject.
- See Also: Related functions that can sometimes be of use, especially when searching for the correct function for our needs.
- Examples: Some examples on how to use the function(s)
—–
Challenge 13
a) Create a sequence of even numbers from 0 to 10 using the seq function
b) - Create an unsorted vector of your favourite numbers.
- Find out how to sort it using
?sort.
- Sort your vector in reverse order.
Getting help on the Web
Usually, your best source of information will be your favorite search engine (Google, Bing, Yahoo, etc.)
Here are some tips on how to use them efficiently:
- Search in English
- Use the keyword “R” at the beginning of your search
- Define precisely what you are looking for
- Learn to read discussion forums. Chances are other people already had the same problem and asked about it. Create your account on forums where questions about R are often asked like stackexchange.
- Don't hesitate to search again with different keywords!
Challenge 14
Find the appropriate functions to perform the following operations
a) Square root
b) Calculate the mean of numbers
c) Combine two dataframes by columns
d) List available objects
Additional resources
Cheat Sheets
https://www.rstudio.com/resources/cheatsheets/
These cheat sheets are also directly available in RStudio (Help → Cheat Sheets)
A few books
- R for Dummies
- A Handbook of Statistical Analyses using R
- R in Action
- Introductory Statistics in R
- A Beginner's Guide to R
- The R Book
A few useful links
R script
Want to revise/practice the material seen here at home?