This is an old revision of the document!
QCBS R Workshops
This series of 10 workshops walks participants through the steps required to use R for a wide array of statistical analyses relevant to research in biology and ecology. These open-access workshops were created by members of the QCBS both for members of the QCBS and the larger community.
Workshop 7: Generalized linear mixed models
Developed by: Cédric Frenette Dussault, Vincent Fugère, Thomas Lamy, Zofia Taranu
Summary: A significant limitation of linear mixed models is that they cannot accommodate response variables that do not have a normal error distribution. Most biological data do not follow the assumption of normality. In this workshop, you will learn how to use generalized linear models, which are important tools to overcome the distributional assumptions of linear models. You will learn the major distributions used depending on the nature of the response variables, the concept of the link function, and how to verify assumptions of such models. We will also build on the previous workshop to combine knowledge on linear mixed models and extend it to generalized linear mixed effect models.
Link to associated Prezi: Prezi
Download the R script and data for this lesson:
- glmm_funs (code to download for use in the GLMM section)
Limitations of linear and linear mixed models
To illustrate why generalized linear models are incredibly useful, it is best to first try to understand the limitations of linear models (workshop 4), which also include linear mixed effect models (workshop 6). Let's load a dataset and try to fit some linear models.
- | loading data
setwd("~/Desktop") mites <- read.csv('mites.csv')
The dataset that you just loaded is a subset of the classic 'Oribatid mite dataset', which has been used in numerous texts (e.g. Borcard, Gillet & Legendre, Numerical Ecology with R), and which is available in the “vegan” library. The dataset consists of 70 moss and mite samples collected at University of Montreal's Station de Biologie des Laurentides, Saint-Hippolyte, QC. Each sample includes measurements for 5 environmental variables and abundance data for 35 mite taxa. In the reduced dataset that we will be using throughout this workshop, we only included the 5 environmental measurements and the abundance of a single mite taxon, “Galumna sp.” Our goal will be to model the abundance, occurrence (presence/absence), and proportion of Galumna as a function of the 5 environmental variables: therefore, we also created a presence/absence variable and a proportion variable for Galumna.
- | exploring the dataset
head(mites) str(mites) # 70 mite communities sampled from moss cores collected at the Station de Biologie des Laurentides, QC. # For each core/sample, the following data is provided: # $Galumna: abundance of mite genus 'Galumna' # $pa: presence (1) or absence (0) of Galumna, irrespective of abundance # $totalabund: total abundance of mites of all species # $prop: proportion of Galumna in the mite community. i.e. Galumna/totalabund # $SubsDens: substrate density # $WatrCont: water content # $Substrate: type of substrate. # $Shrub: abundance of shrubs. Coded as a factor. # $Topo: microtopography. blanket or hummock.
Testing linearity
Can we see any relationship between Galumna and environmental variables?
- | scatterplot matrix
plot(mites)
There seems to be a negative relationship between Galumna abundance and water content (WatrCont). Occurence (presence/absence; pa) and proportion (prop) also seem to be negatively correlated with WatrCont.
We can take a closer look by specifically plotting those 3 response variables as a function of WatrCont:
- | 3 response variables vs. water content
par(mfrow=c(1,3)) #divide plot area in 1 row and 3 columns to have 3 plots in same figure plot(Galumna ~ WatrCont, data = mites, xlab = 'Water content', ylab='Abundance') boxplot(WatrCont ~ pa, data = mites, xlab='Presence/Absence', ylab = 'Water content') plot(prop ~ WatrCont, data = mites, xlab = 'Water content', ylab='Proportion') par(mfrow=c(1,1)) #resets to default setting
Indeed, Galumna seems to vary negatively as a function of WatrCont, i.e. Galumna seems to prefer dryer sites.
Fit linear models (function 'lm') to test whether these relationships are statistically significant.
- | linear models
lm.abund <- lm(Galumna ~ WatrCont, data = mites) summary(lm.abund) lm.pa <- lm(pa ~ WatrCont, data = mites) summary(lm.pa) lm.prop <- lm(prop ~ WatrCont, data = mites) summary(lm.prop)
Yes, there is a strong and significant relationship for all 3 response variables!
Wait a minute… Lets validate these models to make sure that we respect assumptions of linear models, starting with the abundance model.
- | abundance model
plot(Galumna ~ WatrCont, data = mites) abline(lm.abund)
The model does not fit well. It predicts negative abundance values when WatrCont exceeds 600, which does not make any sense. Also, the model does poorly at predicting high abundance values at low values of WatrCont.
- | diagnostic plots
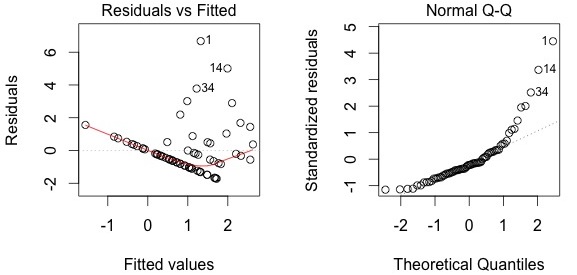
plot(lm.abund)
Diagnostic plots show that the data/model violate assumptions of homogeneity of variance (the graph on the left shows that residuals are larger at higher fitted values) and normality (the graph on the right indicates that residuals are not distributed as expected from a normal distribution, i.e. many points are far from predicted values given by the dotted line). Therefore, we need to reject this model, and can't use it to conclude that Galumna abundance varies as a function of water content. Looking at diagnostic plots for the presence-absence model and the proportion model also indicate that these models are inappropriate:
- | other lm
#Proportion plot(prop ~ WatrCont, data = mites) abline(lm.prop) plot(lm.prop) #Presence/Absence plot(pa ~ WatrCont, data = mites) abline(lm.pa) plot(lm.pa)
It is quite common with biological datasets that assumptions of homogeneity of variance (homoscedasticity) and normality are not met. These two assumptions are the main problem with linear models, and the main reason why we need generalized linear models. Lets revisit the basic equation for a linear model to better understand where these assumptions come from. The equation for a linear model is:
yi = β0 + β1xi + εi, where:
- yi is the predicted value for the response variable,
- β0 is the intercept of the regression line between y and x,
- β1 is the slope of the regression line between y and x,
- xi is the value for the explanatory variable,
- εi are the residuals of the model, which are drawn from a normal distribution with a varying mean but a constant variance.
This last point about εi is important. This is where assumptions of normality and homoscedasticity originate. It means that the residuals (the distance between each observation and the regression line) can be predicted by drawing random values from a normal distribution. Recall that all normal distributions have two parameters, μ (the mean of the distribution) and σ2 (the variance of the distribution):
In a linear model, μ changes based on values of x (the predictor variable), but σ2 has the same value for all values of Y. Indeed, another equation to represent linear models is:
yi ~ N(μ = β0 + β1xi, σ2),
which literally means that any given observation (yi) is drawn from a normal distribution with parameters μ (which depends on the value of xi) and σ2.
Prediction
Predict Galumna abundance at a water content = 300 using this equation and the linear model that we fitted earlier. You will need values for β0 and β1 (regression coefficients) and εi (the deviation of observed values from the regression line)
This model predicts that for a water content value of, say, 300, we should obtain a Galumna abundance of 1.63:
3.44 + (-0.006 x 300) = 1.63.
That is the expected abundance if there was no residual. To get our predicted values, we need to add εi. This is where we use the normal distribution. For x = 300, our model predicts that εi should follow a normal distribution with mean = 1.63. We can find the σ2 value for our abundance model by extracting it from the model summary:
We find that sigma is roughly 1.51. We now have all the coefficients that we need to model Galumna abundance. At a water content of 300, residuals should follow a normal distribution with parameters μ = 1.63 and σ2 = 1.51.
At a water content of 400, residuals εi should follow a normal distribution with parameters μ = 3.44 + (-0.006 x 400) = 1.02 and σ2 = 1.51, etc. Each yi value is modeled using a different normal distribution, with a mean that depends on xi. The variance of all of those normal distributions (σ2), however, is the same. Function lm() finds the optimal σ2 value that minimizes the total residual sum of square and uses it for all normal distributions used to model y. Graphically, this looks like this:
The four normal distributions on this graph represent the probability of observing a given Galumna abundance for 4 different water content values. The mean of the normal distribution varies as a function of water content (hence μ decreases with water content), but σ2 is always 1.51 (i.e. the variance is homogeneous across all values of x). This model is inappropriate for at least two reasons:
1. Values are on average farther from the regression line at low water content values. That is, there is more residual variance around the predicted values for low values of x, such that εi varies as a function of x, thus violating the assumption of homoscedasticity. It makes no sense to use a constant value of σ2: the normal distributions used to predict y at low values of x should ideally be wider (have a larger σ2) than normal distributions used to predict y at large x values, but linear models do not permit this.
2. It makes no sense to use a normal distribution to predict y based on x. Our response variable is abundance, which can only take integer values. Yet, at water content = 300, the abundance value that our model predicts to be the most probable is 1.63! We know that the probability of observing 1.63 individuals at water content = 300 is actually zero, as is the probability of observing any fraction (non-integers). Our predicted values should be modeled using a distribution that only predicts integers, rather than a continuous distribution like the normal distribution. This is a very common problem, as biological data often follows one of the myriad other statistical distributions besides the normal distribution.
Generalized linear models can solve these two problems. Keep reading!
Biological data and distributions
Statisticians have described a multitude of distributions that correspond to different types of data. A distribution provides the probability of observing each possible outcome of an experiment or survey (for example, abundance = 8 Galumna is one such “outcome” of a survey). “Discrete” distributions have a range that only includes integers, while “continuous” distributions have a range that also includes fractions (the normal distribution is an example of a continuous distribution). All distributions have parameters that dictate the shape of the distribution (for example μ and σ2 for the normal distribution). For a good overview of statistical distributions, we recommend that you refer to chapter 4 in Ben Bolker's Ecological Models and Data in R. Here we just discuss briefly a few distributions that are useful for ecologists and generalized linear modeling.
We have already seen that our response variable “Galumna abundance” can only take integer values. Abundance, therefore, follows a discrete distribution, with no fractions in its range. A useful distribution to model abundance data is the “Poisson” distribution, named after Siméon Denis Poisson. The Poisson distribution is a discrete distribution with a single parameter, λ (lambda), which defines both the mean and the variance of the distribution (i.e. the mean and the variance of a Poisson distribution are equal). Here are 3 examples of Poisson distributions with different values of λ, corresponding in this case to the mean number of Galumna observed in a fictive set of samples:
Note that at low λ values (when the mean is close to zero), the distribution is skewed to the right, while at large λ values (large mean) the distribution is symmetrical. The variance increases with the mean, predicted values are always integers, and the range of a Poisson distribution is always strictly positive; all of these properties are useful to model count data, for example abundance of a given taxon, number of seeds in a plot, etc. Our variable mites$Galumna seems to follow a Poisson distribution with a low value of λ (indeed, if we calculate the mean abundance of Galumna across all samples using the function mean(), we find that it is close to zero):
- | histogram for Galumna abundance
hist(mites$Galumna) mean(mites$Galumna)
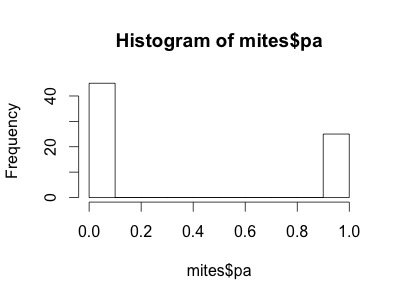
Our variable mites$pa (presence-absence) takes yet another form. It consists of only zeros and ones, such that a Poisson distribution would not be appropriate to model this variable.
- | histogram for Galumna presence-absence
hist(mites$pa)
We need a distribution with a range that only includes two possible outcomes: zero or one. The “Bernoulli” distribution is such a distribution. It is often the first distribution that students of statistics are introduced to, for example when discussing the probability of obtaining the outcome “heads” when flipping a coin. The Bernoulli distribution has only one parameter, p, the probability of success (i.e. the probability of obtaining heads on a coin flip). If we consider that each of our samples is equivalent to a coin toss, then we can use the Bernouilli distribution to calculate the probability of obtaining the outcome “Galumna present” (1) vs. “Galumna absent” (0). Here are some examples of Bernoulli distributions with various probabilities of presence (p):
We can calculate the number of sites where Galumna is present out of the total number of sites to get an idea of what p might be in our case:
- | p for Galumna presence-absence
sum(mites$pa) / nrow(mites)
p for the variable mites$pa is more or less 0.36, such that roughly twice as many sites have the outcome “Galumna absent” (0) than the outcome “Galumna present” (1).
When there are multiple trials/coin tosses, the Bernoulli distribution expands into the binomial distribution, which has the additional parameter n, corresponding to the number of trials. The binomial distribution predicts the probability of observing a given proportion of successes, p, out of a known total number of trials, n. “Successes” can be anything from taxon occurrence, number of surviving individuals out of a sample, etc. Imagine that instead of only working in the Laurentians, we took 50 mite samples at each of 30 regions across Canada. In each sample from each region, we determine if Galumna is present or absent. We could model this data using a binomial distribution with n = 50 samples (i.e. “trials” or coin flips where Galumna can be either present or absent) and p = the average proportion of samples in which Galumna is present. We would have 30 data points, corresponding to the 30 regions. Here are some examples of binomial distributions with n = 50 and 3 different values of p:
Notice that the binomial distribution is right-skewed at low p values but left-skewed at high p values. This is the main difference with the Poisson distribution: the binomial distribution has an upper limit to its range, corresponding to the number of trials, n. Consequently, the binomial distribution is often used to model data where the number of successes are integers and where the number of trials is known. For example, we could use the binomial distribution to model our proportion data, where each individual mite in a sample could be considered a trial, and if the mite is a Galumna individual then the trial is a success. In this case, the number of trials n varies among our 70 samples based on the total number of individuals in the sample, while p, the probability of success, is given by the proportion of Galumna in each sample.
Why are we discussing all of these distributions? Because all of them can be used to replace the normal distribution when calculating predicted values in a linear model. For example, we could use the Poisson distribution and model our abundance data with the following equation:
yi ~ Poisson(λ = β0 + β1xi)
Notice that λ varies as a function of x (water content), meaning that the residual variance will also vary with x. This means that we just relaxed the homogeneity of variance assumption! Also, predicted values will now be integers instead of fractions because they will all be drawn from Poisson distributions with different λ values. The model will never predict negative values because Poisson distributions have strictly positive ranges. By simply switching the distribution of error terms (εi) from normal to Poisson, we solved most of the problems of our abundance linear model. This new model is almost a Poisson generalized linear model, which basically looks like this:
Notice that probabilities of observations/predicted values (in orange, as for the lm model above) are now integers, and that both the variance and the mean of the distribution decline as λ decreases with increasing water content. Why is the fitted line of predicted values curved? Why is this called a “generalized” linear model? Keep reading and you'll find out!
A GLM with binary variables
A common response variable in ecological datasets is the binary variable: we observe a phenomenon X or its “absence”. For example, species presence/absence is frequently recorded in ecological monitoring studies. We usually wish to determine whether a species’ presence is affected by some environmental variables. Other examples include the presence/absence of a disease within a wild population, the success/failure to record a specific behaviour, and the survival/death of organisms. A regression that has a binary response variable is one of many generalized linear models and is called a logistic regression or a logit model.
In R, presence (or success, survival…) is usually coded as 1 and absence (or failure, death…) as 0. A logistic regression (or any other generalized linear model) is performed with the glm() function. This function is different from the basic lm() as it allows one to specify a statistical distribution other than the normal distribution. We've already seen that binary variables are not normally distributed (i.e. we see a peak at 0 and a peak at 1 and nothing in between). Like we have seen in the previous section, the Bernoulli distribution is well suited for binary response variables. The mean of this distribution is the probability p of observing an outcome and the variance is p*(1 - p). The (1 - p) term represents the probability of not observing an outcome. In R, we specify the distribution with the family argument. For the logistic regression, we code it as: family='binomial'. Remember that the Bernoulli distribution is a special case of the binomial distribution when the number of repetitions is 1: R will “understand” that it is a Bernoulli distribution.
When predicting the probability of observing some phenomenon Y, which is a binary variable, the expected values should be bound between 0 and 1: that's the range of a probability value! If we use a basic linear model to relate a binary response variable to various explanatory variables, we might obtain fitted values outside of the [0,1] range, which is nonsensical. The following example will help you understand why a basic linear model is inappropriate here. The next subsection will show you how to avoid this problem with a link function. Briefly, a link function is used to linearize the relationship between predicted values of the response variable and the linear predictor (see next subsection).
- | Inappropriate use of a linear model
model.lm <- lm(pa ~ WatrCont + Topo, data = mites) fitted(model.lm) # The "fitted()" function gives us expected values for the response variable. # Some values are lower than 0, which does not make sense for a logistic regression. # Let’s try the same model with a binomial distribution instead. # Notice the "family" argument to specify the distribution. model.glm <- glm(pa ~ WatrCont + Topo, data = mites, family = binomial) fitted(model.glm) # All values are bound between 0 and 1.
The concept of the link function
To move away from the traditional linear model and to avoid its biases, we need to specify two things when using a logistic regression: a distribution for the residuals of the model and a link function for the expected values. We already presented the Bernoulli distribution in the previous section so let’s have a look at what the link function is.
In the case of a simple linear model of a normally distributed continuous response variable, the following equation gives the expected values:
μ = Xβ
where μ is the expected value of the response variable, X is the model matrix (i.e. representing your data) and β corresponds to the parameters we estimate from the data (i.e. the intercept and the slope). The right-hand side of this equation is called the linear predictor. In mathematical terms, it is the matrix product of the model matrix X of a statistical model and the vector of estimated parameters β. Let’s have a look at this in R:
- | Illustrating the concept of the linear predictor
# Load the CO2 dataset. We used it during workshop 4! data(CO2) head(CO2) # Build a linear model of plant CO2 uptake as a function of CO2 ambient concentration model.CO2 <- lm(uptake ~ conc, data = CO2) # Extract the design matrix of the model with the model.matrix() function. X <- model.matrix(model.CO2) # And the estimated coefficients. B <- model.CO2$coefficients # Let’s multiply both X and B matrices to obtain the linear predictor. # The "%*%" symbol indicates that it is a matrix product. XB <- X %*% B # Compare the values of XB to the values obtained with the predict() function. # All statements should be TRUE. # We use the round() function so that all elements have 5 digits. round(fitted(model.CO2), digits = 5) == round(XB, digits = 5)
When using a simple linear model with a normally distributed response variable, the linear predictor is directly equal to the expected values of the model. But, what if our response variable is not normally distributed? If that is the case, we have to use a transformation on the expected values, i.e. a link function. A link function can be understood as a transformation of the expected values so that it can be linearly related to the linear predictor:
g(μ) = Xβ
where g(μ) is the link function for the expected values. This allows us to relax the normality assumption. In the case of a binary response variable, the link function is called the logit function and is given by:
logit(μ) = log (μ / 1-μ) = Xβ
where μ represents expected values (i.e. the probability that Y = 1 because we observed the presence of a species, disease, success, or some other event). The ratio μ / 1-μ represents the odds that some outcome occured and it transforms the expected values into continuous values from 0 to infinity. If we have a 0.8 probability of observing species X, then our odds are 4 times more likely to observe the species than to not observe it: 0.8/(1-0.8) = 4. The log transformation, called the log odds, allows values to be spread across -infinity to infinity. Hence, the logit function took the expected values of a model and transformed them into continuous values without boundaries. The expected values can now be directly related to a linear predictor. This is why we still call this model a generalized linear model even though the plot of our response variable as a function of some explanatory variable doesn't look like a “straight line”!
- | A simple example for logistic regression
# Let’s build a regression model of the presence/absence of a mite species (Galumna sp.) # as a function of water content and topography. # To do this, we need to use the glm() function and specify the family argument. logit.reg <- glm(pa ~ WatrCont + Topo, data = mites, family = binomial(link = "logit")) # The logit function is the default for the binomial distribution, # so it is not necessary to include it in the "family" argument: logit.reg <- glm(pa ~ WatrCont + Topo, data = mites, family = binomial) summary(logit.reg)
CHALLENGE 4
Using the bacteria dataset (from the MASS package), model the presence of H. influenzae as a function of treatment and week of test. Start with a full model and reduce it to the most parsimonious model.
Interpreting the output of a logistic regression
The output of the previous logistic regression indicates that both water content and topography are significant, but how do we interpret the slope coefficients? Remember that we applied a transformation on our expected values (i.e. the probability that Y = 1) so we have to use a reverse function to properly interpret the results. We can use the natural exponential function ex to obtain the odds of probability of success for each explanatory variable.
- | Interpreting the output of a logistic regression
# Obtaining the odds of the slope. # Use the "exp()" function to put the coefficients back on the odds scale. # Mathematically, this line of code corresponds to: # exp(model coefficients) = exp(log(μ / (1 - μ)) = u / (1 - μ) # This corresponds to an odds ratio! exp(logit.reg$coefficients[2:3]) # WatrCont TopoHummock # 0.9843118 8.0910340 # To obtain confidence intervals on the odds scale: exp(confint(logit.reg)[2:3,]) # 2.5 % 97.5 % # WatrCont 0.9741887 0.9919435 # TopoHummock 2.0460547 38.6419693
Note that the odds values here are considered when all other parameters are kept constant. The topography parameter value is 8.09. It means that the probability of observing Galumna sp. is 8.09 times more likely when the topography is hummock compared to blanket.
When the odds value is smaller than 1, interpretation is a little bit more complicated. When this is the case, we have to take the inverse value (i.e. 1 divided by the odds) to facilitate interpretation. The interpretation is then how LESS likely it is to observe the event of interest. For water content, the odds is 0.984. The inverse is 1 / 0.984 = 1.0159. This means that a one-unit increase in water content decreases the likelihood of observing Galumna sp. by 1.0159. We can also substract 1 from the odds value to obtain a percentage: (1.0159 - 1) * 100 = 1.59% decrease in probability of observing Galumna sp. with a one-unit increase in water content. To convince ourselves that it is an appropriate interpretation, we can plot the presence of Galumna sp. as a function of water content. We see that, on average, Galumna sp. presence is higher at lower water content than its “absence”.
When the parameter estimate is between 0 and 1 on the odds scales, it indicates a negative relationship between the response variable and the explanatory variable. If the parameter is greater than 1, it indicates a positive relationship between the response variable and the explanatory variable. If the confidence interval includes 1, it indicates that the variable is not significant. Remember that a value of 1 on the odds scale means that the probability of Y = 1 is the same as the probability of Y = 0 (i.e. when p = 0.5, 0.5/(1-0.5) = 1).
If you want to obtain the probabilities instead of the odds for each explanatory variable, the inverse logit function is what you need:
logit-1 = 1/(1+1/exp(x))
where x is the parameter to transform from log odds to the probability scale. The parameter estimate of topography in our logit.reg model is 2.091, which is on the log odds scale. So, the probability value is given by:
1/(1+1/exp(2.091)) = 0.89 which is the same as 1/(1+1/8.09). Remember that the value 8.09 is on the odds scale. We have a 0.89 probability of observing Galumna sp. when the topography is Hummock.
- | Some extra math about the inverse logit
# Let's start with our odds ratio for topography from the logit.reg model: µ/ (1 - µ) = 8.09 # Let's rearrange this to isolate µ µ = 8.09(1 - µ) = 8.09 - 8.09µ 8.09µ + µ = 8.09 µ(8.09 + 1) = 8.09 µ = 8.09 / (8.09 + 1) µ = 1 / (1 + (1 / 8.09)) = 0.89 # We obtained the same result without using the exp() function!
Predictive power and goodness-of-fit assessment
An easy and intuitive way to evaluate the predictive power of your model is to compare its deviance to the deviance of a null model. Deviance can be understood as a generalisation of the residual sum of squares when models are estimated by maximum likelihood (i.e. it is how parameters are estimated in GLM). This allows us to compute a pseudo-R2 statistic, which is analogous to the coefficient of determination R2 in ordinary least square regression (i.e. the basic method for linear models). The null model is a model without any explanatory variable. Its notation in R is: null.model <- glm(Response.variable ~ 1, family = binomial). The generic formula to compute a pseudo-R2 is given by:
Pseudo-R2 = (null deviance – residual deviance) / null deviance
where “null deviance” is the deviance of the null model and “residual deviance” is the deviance of the model of interest. The difference is divided by the null deviance so that the result is bound between 0 and 1.
- | Pseudo-R2 of a logistic regression
# Residual and null deviances are already stored in the glm object. objects(logit.reg) pseudoR2 <- (logit.reg$null.deviance – logit.reg$deviance) / logit.reg$null.deviance pseudoR2 # [1] 0.4655937
Hence, the model explains 46.6% of the variability in the data.
Recently, Tjur (2009) proposed a new statistic, the coefficient of discrimination (D), to evaluate the predictive power of logistic regression models. Intuitively, D is a measure of how well a logistic regression can classify an outcome as a success or a failure. In mathematical terms, it is the difference between the means of expected probability values for successes (i.e. Y =1) and failures (i.e. Y = 0):
where is the mean of expected probability values when the outcome is observed and
is the mean of expected probability values when the outcome is not observed. A D value close to 1 indicates that the model gives a high probability of observing an outcome to cases where the outcome was actually observed and a low probability of observing an outcome to cases where the outcome was not observed. A D value close to 0 indicates that the model is not efficient at discriminating between the occurrences and “non occurrences” of an outcome. The following code shows how to obtain D and how to plot the histograms of
and
.
- | The coefficient of discrimination and its visualisation
install.packages("binomTools") library("binomTools") # The Rsq function computes several fit indices, # including the coefficient of discrimination. # For information on the other fit indices, see Tjur (2009). # The plot shows the distribution of expected values when the outcome is observed # and not observed. # Ideally, the overlap between the two histograms should be small. fit <- Rsq(object = logit.reg) fit # R-square measures and the coefficient of discrimination, 'D': # # R2mod R2res R2cor D # 0.5205221 0.5024101 0.5025676 0.5114661 # # Number of binomial observations: 70 # Number of binary observation: 70 # Average group size: 1 plot(fit, which = "hist")
To assess the goodness-of-fit of a logistic regression, the diagnostic plots (see workshop 4) are not useful. Instead, you can do a Hosmer-Lemeshow test to evaluate whether your model is appropriate. This test separates the expected values (ordered from smallest to largest) in groups of approximately equal size. Ten is usually the recommended group number. In each group, we compare the observed and expected number of outcomes. It is similar to a chi-square test with G - 2 degrees of freedom (G is the number of groups). In R, this test is available in the binomTools package.
- | The Hosmer-Lemeshow test
fit <- Rsq(object = logit.reg) HLtest(object = fit) # The p value is 0.9051814. Hence, we do not reject the model. # We can consider it as appropriate for the data.
CHALLENGE 5
Assess goodness-of-fit and predictive power of the model.bact2 model.
How can you improve the predictive power of this model?
Plotting results
Once we’ve created a model and verified its validity, we can plot the results to show how the response variable is related to some explanatory variables. One way to graphically summarise the data is by plotting both the response variable and the expected values as a function of some predictor. Here is an example with the ggplot2 package. See workshop 3 for more information on this package.
- | Plotting the results of a logistic regression
library(ggplot2) ggplot(mites, aes(x = WatrCont, y = pa)) + geom_point() + stat_smooth(method = "glm", family= "binomial", se = FALSE) + xlab("Water content") + ylab("Probability of presence") + ggtitle("Probability of presence of Galumna sp. as a function of water content")
An example with proportion data
Proportion data are more similar to binary data than you might think! Suppose you’re in the field to estimate the prevalence of a disease within wild populations of white-tailed deer along an environmental gradient of resource availability. You sample ten individuals in ten different populations to have an estimate of disease prevalence. Your data sheet has ten lines with the following information: population ID and number of individuals with the disease. At first sight, it seems you have count data, but this is inaccurate because you know how many individuals were sampled in each population. In fact, you have proportion data: it is the number of individuals with the disease out of ten sampled individuals. If you remember what you've read in the section on distributions, you should choose a binomial distribution to model these data. Let’s illustrate this with an example:
- | GLM and proportion data
# Let’s generate some data based on the deer example: # We randomly choose a number between 1 and 10 for the number of infected deer. # Ten deers were sampled in ten populations. # Resource availability is an index to characterise the habitat. set.seed(123) n.infected <- sample(x = 1:10, size = 10, replace = TRUE) n.total <- rep(x = 10, times = 10) res.avail <- rnorm(n = 10, mean = 10, sd = 1) # Next, let’s build the model. Notice how the proportion data is specified. # We have to specify the number of cases where disease was detected # and the number of cases where the disease was not detected. prop.reg <- glm(cbind(n.infected, n.total - n.infected) ~ res.avail, family = binomial) summary(prop.reg) # If your data is directly transformed into proportions, here is the way to do it in R: # Let's first create a vector of proportions prop.infected <- n.infected / n.total # We have to specify the "weights" argument in the glm function to indicate the number of trials per site prop.reg2 <- glm(prop.infected ~ res.avail, family = binomial, weights = n.total) summary(prop.reg2) # The summaries of both prop.reg and prop.reg2 are identical!
GLMs with count data
To illustrate count data we will use a new dataset called faramea.
- | loading data
faramea <- read.csv(‘faramea.csv’, header = TRUE)
In this dataset, the number of trees of the species Faramea occidentalis was measured in 43 quadrats in Barro Colorada Island in Panama. For each quadrat, environmental characteristics were also recorded such as elevation or precipitation. Let’s take a look at the number of Faramea occidentalis found at each quadrat.
- | plot the data
hist(faramea$Faramea.occidentalis, breaks=seq(0,45,1), xlab=expression(paste("Number of ", italic(Faramea~occidentalis))), ylab="Frequency", main="", col="grey")
We can see that there are only positive and integer values. Given these specificities, the Poisson distribution, described above, seems to be the perfect choice to model this data.
Poisson GLMs
As we saw above, the Poisson distribution is particularly relevant to model count data because it:
- specifies the probably only for integer values
- P(y<0) = 0, hence the probability of any negative value is null
- the mean-variance relationship allows for heterogeneity (e.g. when variance generally increases with the mean)
In this example, we want to test whether elevation (a continuous explanatory variable) influences Faramea occidentalis abundance. Hence, a Poisson GLM (i.e. a simple Poisson regression) seems to be a good choice to model the number of Faramea occidentalis as a function of elevation. Poisson GLMs are usually a good way to start modeling count data.
But what does a Poisson GLM do?
It assumes that the response variables yi have been generated by a Poisson distribution with mean and variance µi.
Yi ∼ Poisson(µi)
with E(Yi) = Var(Yi) = µi
Recall from above that µi can be replaced the systematic model
Yi ~ Poisson(β0 + β1Xi)
A systematic part is used as a linear combination of unknown parameters β representing the effects of different explanatory variables. X is the covariate matrix (which does not include an intercept term in this example). This systemtatic part is define as:
β0 + Xi.β
The mean of the distribution µi is related to the systematic part using a logarithm link function. As a result, the relationship between the mean and the linear predictor is log-linear.
- log(µi) = β0 + Xi.β
or
- µi = exp(β0 + Xi.β)
The Poisson distribution gives you the probability that a particular Yi value is observed for a given mean µi = exp(β0 + Xi.β). In this model, the unknown parameters are included in the vector of regression coefficients β (plus the intercept β0) and can be estimated using maximum-likelihood (ML) estimation.
Fitting a Poisson GLM in R requires only setting family = poisson in the function glm(). By default the link function is log.
- | fit a Poisson GLM
glm.poisson = glm(Faramea.occidentalis~Elevation, data=faramea, family=poisson) summary(glm.poisson)
The output is similar to a 'lm' output (see workshop 4) and gives you the parameter estimates which can also be retrieved using other functions:
- | parameter estimates
# intercept summary(glm.poisson)$coefficients[1,1] # slope of elevation summary(glm.poisson)$coefficients[2,1]
Model validation and the problem of overdispersion
An important aspect of the summary can be found in the last lines.
- | Poisson GLM summary
summary(glm.poisson) # Null deviance: 414.81 on 42 degrees of freedom # Residual deviance: 388.12 on 41 degrees of freedom
Remember that ML estimation is used to estimate the parameters. In the goodness-of-fit section we mentioned that the deviance was a ML equivalent of the sum of squares in linear models. Here, the null deviance and the residual deviance are equivalent to the total sum of squares and the residual sum of squares respectively. The residual deviance is defined as twice the difference between the log likelihood of a model that provides a perfect fit to the data (a saturated model) and the log likelihood of the model. If our model is correct the asymptotic distribution of the residual deviance is approximated using χ² distribution with n-p-1 degrees of freedom (computed as n-p-1, where n is the number of observations and p the number of covariates). This implies that residual deviance should be equal to the residual degrees of freedom. In our example, the residual deviance equals 388.12 while we have 41 (43-1-1) degrees of freedom. This former is greater to the former by 9.5 times, the model is then overdispersed.
Overdispersion As a consequence overdispersion can be computed for any model using the parameter φ:
φ = residual deviance / residual degrees of freedom * φ < 1 indicates underdispersion * φ = 1 indicates no overdispersion * φ > 1 indicates overdispersion
Why does a Poisson GLM exhibit overdispersion? This arises when the variance of the data is higher than expected from the Poisson distribution. This frequently occurs when data includes many zeros and/or many very high values. Looking back at the distribution of our data (above) suggests that our data contains many zero preventing us to use a Poisson GLM. Overdispersion may also result from missing covariates, missing interaction terms or presence of strong outliers, preventing us from using a Poisson GLM.
The Poisson distribution can account only partially for heterogeneity in the data due to the mean variance relationship, but in some cases variance increases even higher than the mean. Computing the mean and the variance of our dataset suggests this is occurring:
- | mean and variance of the data
mean(faramea$Faramea.occidentalis) var(faramea$Faramea.occidentalis)
In practice, Poissons GLM are useful for describing the mean µi but underestimates the variance in the data, making all model-based tests too liberal! There are two ways of dealing with overdispersion and will be developed below:
- correct for it by doing a quasi-Poisson GLM
- choose another distribution such as the negative binomial
quasi-Poisson GLMs
The principle behind a quasi-Poisson GLM is very simple; the overdispersion parameter (φ) is added to the expected variance equation:
E(Yi) = µi
Var(Yi) = φ.µi
The systematic part and the link function remains the same. The difference is that φ will first be estimated to correct the model. Parameter Estimates will be the same but the standard errors of the parameters are multiplied by √φ, in other terms, some marginally significant p-values may no longer hold.
In R, The 'quasipoisson' family object can be used to deal with count data exhibiting overdispersion (the 'quasibinomial' family object can do the same for binomial data). The fitted φ value will be returned in the summary of the GLM. There are two ways to perform this GLM:
- | fit a new quasi-Poisson GLM
# Option 1, fit a new quasi-Poisson GLM glm.quasipoisson = glm(Faramea.occidentalis~Elevation, data=faramea, family=quasipoisson) # Option 2, build from the previous model and update it: glm.quasipoisson = update(glm.poisson,family=quasipoisson) # output summary(glm.quasipoisson)
If you look at the summary of the model you will see that φ is estimated as 15.97. We then made a good choice by updating the model to account for overdispersion. However if we look at P-values we can note that elevation is no longer significant. Yet, 15.97 is quite a lot of overdispersion, and in general quasi-Poisson GLMs will be favoured when φ is included between 1 and 15 though these limits are arbitrary. When overdispersion is higher than 15-20 we recommend moving to the negative binomial. For the sake of pedagogy, we will consider that we are not happy with this model to fit a final negative-binomial GLM to our data.
Two other points are important to keep in mind when using quasi-Poisson GLMs and dealing with overdispersion:
- quasi-Poisson GLMs do not have AIC scores. An important aspect is that quasi-Poisson GLMs do not correspond to models with fully specified likelihoods and rely on quasi-ML estimation (i.e. pseudolikelihood). One consequence is that quasi-Poisson GLMs do not have AIC scores for model comparisons. However, variants of AIC have been developed to deal with this situation (e.g. quasi-AIC).
- Overdispersion affects model comparison. Indeed overdispersion also influences the comparison of two nested models and has to be taken into account when φ is known. For instance, let’s assume that we want to compare GLM1, with p1 parameters to GLM2, with p2 parameters, such that GLM1 is nested within GLM2 and p2 > p1. Model comparison is achieved based on a generalized likelihood ratio test, which can be written as a function of the difference of deviances between the two GLMs, D1 and D2 respectively. If Overdispersion is known, deviances have to be scaled (i.e. corrected) accordingly as D* = D/φ. The final test will be based on the criterion D1* - D*2 which is assumed to follow a χ² distribution with p2-p1 degrees of freedom when GLM1 is correct.
- In some cases φ is not known. For instance, this occurs when you run a GLM with a normal error distribution. In that case, φ can be estimated a posteriori using the residual deviance of the larger model so the criterion becomes [(D1-D2)/(p2-p1)]/[D2/(n-p2)] and is assumed to follow a F distribution with p2-p1 and n-p2 degrees of freedom.
Negative binomial GLMs
GLM with a negative binomial (NB) distribution are favored when overdispersion is extreme. The NB distribution contains an additional parameter k, particularly handy for count data containing a preponderance of zeros. Before we go into R stuff, we should see what lies behind a negative binomial GLM. A NB distribution is actually a combination of two distributions: a Poisson distribution and a gamma distribution. The NB distribution first assumes that a discrete random variable is Poisson distributed but its mean, µ is assumed to follow a gamma distribution. The mixture between the Poisson and gamma distributions can be simplified into a density function specific to the NB which has two parameters µ and k.
Y ~ NB(µ, k)
E(Y) = µ and Var(Y) = µ + µ²/k
Here, we can see how overdispersion will be accounted for by NB distribution in GLMs. The second term of the variance determines the amount of overdispersion. In fact, it is indirectly determined by k, where k is also called the dispersion parameter. If k is large (relative to μ²), the second term, µ²/k approximates 0, and the variance of Y is μ. In such cases the NB converges to the Poisson distribution and you might as well use a Poisson distribution. The smaller k, the larger the overdispersion. Just like with others GLMs, a NB GLM is specified following the fundamental three steps. It first assumes that Yi is negative binomial distributed with mean μi and parameter k.
Yi ∼ NB(µi, k)
E(Yi) = µi and Var(Yi) = µi + µi²/k
The two last steps define the systematic part and the link function between the mean of Yi and the predictor function. In NB GLMs the link function is logarithmic ensuring that fitted values are always positive.
- log(µi) = β0 + Xi.β
or
- µi = exp(β0 + Xi.β)
NB is not in the glm() function so you need to install and load the MASS library. You do not remember if you already installed this package? No problem, you can use the following function:
- | Is 'Mass' installed?
ifelse(length(which(installed.packages() == "MASS")) == 0, {print("MASS not installed. Installing... "); install.packages("MASS")}, print("MASS already installed"))
Alternatively, if you know that this package is not installed you can directly use the command
- | install 'MASS'
install.packages("MASS")
and remember to load the package
- | load 'MASS'
library("MASS")
The negative binomial GLM can be build using the glm.nb() function:
- | fit a NB GLM'
glm.negbin = glm.nb(Faramea.occidentalis~Elevation, data=faramea) summary(glm.negbin)
The summary is similar to other GLMs summaries (e.g. Poisson GLMs), though we now have a parameter theta, which stands for parameter k in the variance of the NB distribution. Its standard error is also provided, but care is needed with its use as the interval is not symmetric and we are testing on the boundary.
Plotting the final GLM to the data
The NB GLMs appear to be the best fit to our data. We can plot the relationship between the abundance of Faramea occidentalis and elevation.
- | plot A GLM'
# plot the observed data plot(faramea$Elevation, faramea$Faramea.occidentalis, xlab="Elevation (m)", ylab=expression(paste("Number of", " ", italic(Faramea~occidentalis))), pch=16, col=rgb(4,139,154,150,maxColorValue=255)) # pull values for intercept and beta from the summary and put them in the exponential equation curve(exp(summary(glm.negbin)$coefficients[1,1]+summary(glm.negbin)$coefficients[2,1]*x),from=range(faramea$Elevation)[1],to=range(faramea$Elevation)[2],add=T, lwd=2, col="orangered") # pull the standard error as well to plot the equations for confidence envelope curve(exp(summary(glm.negbin)$coefficients[1,1]+1.96*summary(glm.negbin)$coefficients[1,2]+summary(glm.negbin)$coefficients[2,1]*x+1.96*summary(glm.negbin)$coefficients[2,2]),from=range(faramea$Elevation)[1],to=range(faramea$Elevation)[2],add=T,lty=2, col="orangered") curve(exp(summary(glm.negbin)$coefficients[1,1]-1.96*summary(glm.negbin)$coefficients[1,2]+summary(glm.negbin)$coefficients[2,1]*x-1.96*summary(glm.negbin)$coefficients[2,2]),from=range(faramea$Elevation)[1],to=range(faramea$Elevation)[2],add=T,lty=2, col="orangered")
We can see that the number of Faramea occidentalis significantly decreases with elevation. However, the confidence envelope of the NB model is large at low elevation.
Conclusion on GLMs with count data
All the GLMs introduced (Poisson, quasi-Poisson and NB) to model count data use the same log-linear mean function (log(µ) = X.β), but make different assumptions about the remaining likelihood. Quasi-Poisson and NB are favored to deal with overdispersion. However, in some cases the data may contains too many zeros and zero-augmented models can be useful as they extend the mean function by modifying (typically, increasing) the likelihood of zero counts (e.g. zero-inflated Poisson [ZIP]).
Generalized linear mixed models (GLMMs)
As seen above, heterogeneous response distributions can be accurately modelled by relaxing the assumptions of normally distributed errors and letting the errors take on different distribution families (e.g., Poisson or negative binomial). What happens if there is a structure in the dataset such that observations sampled in certain sites are more similar (correlated) than observations sampled in different sites? As we saw in Workshop 6, we can account for this lack of independence by adding random effects in the model. Recall that intercepts and/or slopes are allowed to vary among grouping factors (random effects) and the variability in slopes/ intercepts among groups is assumed to follow a normal distribution. Random effects are also referred to as shrinkage estimates because they represent a weighted average of the group-level and the overall fit (fixed effect). The amount of shrinking towards the overall fit is more severe if the within-group variability is large compared to the among-group variability (i.e., when we have less confidence in the random estimate due to low sample size or high within-group variability, the estimate is 'pulled' towards the fixed effect).
The properties of Linear Mixed Models (Workshop 5) which incorporate random effects and those of Generalized Linear Models (seen above) which handle non-normal data can be combined to effectively model such instances in a Generalized Linear Mixed Model framework. The extension of GLMs to account for this additional structure follows similar steps introduced in the mixed modelling workshop.
To provide a brief overview of GLMMs, we will visit a case study presented by Bolker et al. (2009) and later by Bolker et al. (2011) where the structure (or lack of independence) in the dataset was due to observations being measured across different populations. The authors evaluated the extent to which the response of Arabidopsis thaliana (mouse-ear cress) to its environmental drivers (nutrient availability and herbivory) varied as a result of the population and/or genotype they belonged to.
Using the Arabidopsis dataset and detailed analysis presented by Bolker and colleagues, we will explore the eff ects of nutrient levels, herbivory and their interaction ( fixed effects) on the production Arabidopsis thaliana and the across-population and/or across-genotype variability in these relationships (random eff ects).
- | Loading
# First load and view the dataset dat.tf <- read.csv("Banta_TotalFruits.csv") str(dat.tf) # X observation number # reg factor for one of three regions; Netherlands, Spain or Sweden # popu factor with a level for each population (random effect) # gen factor with a level for each genotype (random effect) # rack nuisance factor for one of two greenhouse racks # nutrient factor with levels for low (value = 1) or high (value = 8) nutrients (fixed effect) # amd factor with levels for no damage or simulated herbivory (apical meristem damage) (fixed effect) # status nuisance factor for germination method # total.fruits the response variable; an integer indicating the number of fruits per plant # 2-3 genotypes nested within each of the 9 populations table(dat.tf$popu,dat.tf$gen) # Housekeeping: make integers into factors, relevel clipping (amd) and rename nutrient levels dat.tf <- transform(dat.tf, X=factor(X), gen=factor(gen), rack=factor(rack), amd=factor(amd,levels=c("unclipped","clipped")), nutrient=factor(nutrient,label=c("Low","High"))) # Install/load packages if(!require(lme4)){install.packages("lme4")} require(lme4) if(!require(coefplot2)){install.packages("coefplot2",repos="http://www.math.mcmaster.ca/bolker/R",type="source")} require(coefplot2) if(!require(reshape)){install.packages("reshape")} require(reshape) if(!require(ggplot2)){install.packages("ggplot2")} require(ggplot2) if(!require(plyr)){install.packages("plyr")} require(plyr) if(!require(gridExtra)){install.packages("gridExtra")} require(gridExtra) if(!require(emdbook)){install.packages("emdbook")} require(emdbook) source("glmm_funs.R")
Structure in dataset
When we look at the interaction between nutrient and clipping across the 9 different populations, we note that there is a consistent nutrient e ffect (higher baseline under the High nutrient treatment). The effect of clipping is weaker, but in some populations we note a negative slope.
- facet by popu
ggplot(dat.tf,aes(x=amd,y=log(total.fruits+1),colour=nutrient)) + geom_point() + stat_summary(aes(x= as.numeric(amd)),fun.y=mean,geom="line") + theme_bw() + theme(panel.margin=unit(0,"lines")) + scale_color_manual(values=c("#3B9AB2","#F21A00")) + # from Wes Anderson Zissou palette facet_wrap(~popu)
A similar plot can be made for the 24 different genotypes (changing facet_wrap(~gen)).
Choosing an error distribution
The response variable is count data which suggests that a Poisson distribution should be used. Recall that an important property of the Poisson distribution is that the variance is equal to the mean. However, as we will see below, the group variances increase with the mean much more rapidly than expected under the Poisson distribution.
- | Heterogeneity across groups
# Create new variables that represents every combination nutrient x clipping x random factor dat.tf <- within(dat.tf, { # genotype x nutrient x clipping gna <- interaction(gen,nutrient,amd) gna <- reorder(gna, total.fruits, mean) # population x nutrient x clipping pna <- interaction(popu,nutrient,amd) pna <- reorder(pna, total.fruits, mean) }) # Boxplot of total fruits vs new variable (genotype x nutrient x clipping) ggplot(data = dat.tf, aes(factor(x = gna),y = log(total.fruits + 1))) + geom_boxplot(colour = "skyblue2", outlier.shape = 21, outlier.colour = "skyblue2") + theme_bw() + theme(axis.text.x=element_text(angle=90)) + stat_summary(fun.y=mean, geom="point", colour = "red") # Boxplot of total fruits vs new variable (population x nutrient x clipping) ggplot(data = dat.tf, aes(factor(x = pna),y = log(total.fruits + 1))) + geom_boxplot(colour = "skyblue2", outlier.shape = 21, outlier.colour = "skyblue2") + theme_bw() + theme(axis.text.x=element_text(angle=90)) + stat_summary(fun.y=mean, geom="point", colour = "red")
As illustrated above, there is a large amount of heterogeneity among the group variances even when the response variable is transformed. We also note that some groups have a mean and variance of zero.
To determine which distribution family to use, we can run a diagnostic plot of the group variances vs their respective means. We provide an example below for the genotype x nutrient x clipping grouping.
- If we observe a linear relationship between the variance and the mean with a slope = 1, then the Poisson family is appropriate,
- If we observe a linear mean-variance relationship with a slope > 1 (i.e. Var = φµ where φ > 1), then the quasi-Poisson family (as introduced above) should be applied,
- Finally, a quadratic relationship between the variance and the mean (i.e. Var = µ(1 + α
) or µ(1 + µ/k)), is characteristic of overdispersed data that is driven by an underlying heterogeneity among samples. In this case, the negative binomial (Poisson-gamma) would be more appropriate.

From the plot above we note that a linear quasi-Poisson may be better than the negative binomial, but additional modelling is needed.
Poisson GLMM
Let's build a GLMM using the glmer function of the lme4 package. This model has a random intercept for all genotype and population treatments. We include the nuisance variables (rack and germination method) as fixed effects. Given the mean-variance relationship from above, we most likely will need a model with overdispersion, but let's start with a Poisson model.
- | Poisson GLMM
mp1 <- glmer(total.fruits ~ nutrient*amd + rack + status + (1|popu)+ (1|gen), data=dat.tf, family="poisson")
We can check for overdispersion using a function overdisp_fun provided by Bolker et al. (2011) which divides the Pearson residuals by the residual degrees of freedom and tests whether these values differ significantly (i.e., whether the ratio of residual deviance to residual df is significantly different from 1). A low p-value indicates that the data are over dispersed (i.e., ratio is significantly different from 1).
- | Overdispersion check
# Overdispersion? overdisp_fun(mp1) # Or as above, we can approximate this by dividing the residual deviance by the residual df summary(mp1) # residual deviance = 18253.7 and resid df = 616
The ratio is significantly greater than unity, so as expected, we need to try a different distribution where the variance increases more rapidly than the mean, such as the negative binomial.
Negative binomial (Poisson-gamma) GLMM
Recall that the negative binomial (or Poisson-gamma) distribution meets the assumption that the variance is proportional to the square of the mean.
- | NB
# Note: This model converges well if you are running R version 3.0.2, but may not converge with later # versions. If you are having convergence issues, please try with version 3.0.2. mnb1 <- glmer.nb(total.fruits ~ nutrient*amd + rack + status + (1|popu)+ (1|gen), data=dat.tf, control=glmerControl(optimizer="bobyqa")) # Although beyond the scope of this workshop, the new "control" argument specifies the way we # optimize the parameter values (i.e. by taking the derivative of a function or proceeding by iteration). # When taking the derivative is not possible, an iterative algorithm such as bobyqa (Bound Optimization # BY Quadratic Approximation) is used. # Overdispersion? overdisp_fun(mnb1)
The ratio is now much closer to unity (although the p-value is still less than 0.05).
Poisson-lognormal GLMM
Another option that we have not yet seen is to use a Poisson-lognormal distribution. This approach deals with overdispersion by implementing an observation-level random effects (OLRE; see Harrison (2014)), which model the extra-Poisson variation in the response variable using a random effect with a unique level for every data point.
A Poisson-lognormal model effectively places a lognormal prior on εi. A Poisson-lognormal distribution with mean µ and lognormal prior variance σ2 has variance:
var(y) = µ + µ2 [exp(σ2) - 1]
In contrast, for the negative binomial, we saw that the distribution was given by:
var(y) = µ + µ2/k
More generally, the variance term σ2 in the Poisson-lognormal distribution will depend on the grouping level we select (e.g., at the individual, genotype or population level). That is, the Poisson-lognormal model can allow for a more flexible approach to assigning the observed aggregation to different sources of heterogeneity. Here, to implement the observation-level random effect, we will evaluate it at the individual level.
- | PL GLMM
mpl1 <- glmer(total.fruits ~ nutrient*amd + rack + status + (1|X) + (1|popu)+ (1|gen), data=dat.tf, family="poisson", control=glmerControl(optimizer="bobyqa")) overdisp_fun(mpl1)
We did it! The ratio between residual deviance and residual df now meets our criterion (in fact, it is even smaller 1).
Random intercepts
Now that we have the appropriate error distribution, we can test the importance of the random intercepts (for population and genotype) by comparing nested models with and without the random effects of interest using either:
- an information theoretic approach (such as, Akaike Information Criterion; AIC), which as we saw in Workshop 6 examines several competing hypotheses (models) simultaneously to identify the model with the highest predictive power given the data. As before, we will use the AICc to correct for small sample sizes. Or,
- a frequentist approach (traditional null hypothesis testing or drop1 approach), where the significance of each term is evaluated in turn (by comparing nested models together using the
anova()function and the significance of the likelihood ratio test; LRT). It's important to keep in mind that with this approach we are testing a null hypothesis of zero variance for the random effects, but given that we cannot have negative variance, we are testing the parameter on the boundary of its feasible region. Therefore, the reported p value is approximately twice what it should be (i.e., we've truncated half of the possible values that fall below 0).
- Evaluating the random intercepts
summary(mpl1)$varcor # popu only mpl1.popu <- glmer(total.fruits ~ nutrient*amd + rack + status + (1|X) + (1|popu), data=dat.tf, family="poisson", control=glmerControl(optimizer="bobyqa")) # gen only mpl1.gen <-glmer(total.fruits ~ nutrient*amd + rack + status + (1|X) + (1|gen), data=dat.tf, family="poisson", control=glmerControl(optimizer="bobyqa")) # IC approach using AICc ICtab(mpl1, mpl1.popu, mpl1.gen, type = c("AICc")) # dAICc df # mpl1 0.0 10 # mpl1.popu 2.0 9 # mpl1.gen 16.1 9 # Frequentist approach using LRT anova(mpl1,mpl1.popu) # Data: dat.tf # Df AIC BIC logLik deviance Chisq Chi Df Pr(>Chisq) # mpl1.popu 9 5017.4 5057.4 -2499.7 4999.4 # mpl1 10 5015.4 5059.8 -2497.7 4995.4 4.0639 1 0.04381 * # --- # Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 anova(mpl1,mpl1.gen) # Df AIC BIC logLik deviance Chisq Chi Df Pr(>Chisq) # mpl1.gen 9 5031.5 5071.5 -2506.8 5013.5 # mpl1 10 5015.4 5059.8 -2497.7 4995.4 18.177 1 2.014e-05 *** # --- # Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
The model without the random intercept for genotype is within 2 AICc units of the full model, which indicates that they are equally plausible (i.e., little evidence for including a random intercept for genotype). However, when using the Likelihood approach, and keeping in mind the boundary effect (p-values are inflated by a factor of 2), we note that p « 0.05 in both anova tests. Thus the model with both random terms (mpl1) is selected.
Parameter plots
A graphical representation of the model parameters can be obtained using the coefplot2 function. For example, to view the variance terms of our three random intercepts:
- Coefplot2
# Variance terms coefplot2(mpl1,ptype="vcov",intercept=TRUE,main="Random effect variance") # Fixed effects coefplot2(mpl1,intercept=TRUE,main="Fixed effect coefficient")
Note that the error bars are only shown for the fixed effects because glmer doesn't give us information on the uncertainty of the variance terms.
Random intercept plots
We can also extract the random effect (or group-level) deviations from the fixed intercept using the ranef function. This will tell us how much the intercept is shifted up or down in particular populations or genotypes relative to the fixed intercept. The deviations can then be plotted using dotplot, which will return a two-facetted plot for each random effect (i.e., popu and gen). Note: the grid.arrange function was used to omit the observation-level random effect (i.e. (1|X)).
- dotplot
pp <- list(layout.widths=list(left.padding=0, right.padding=0), layout.heights=list(top.padding=0, bottom.padding=0)) r2 <- ranef(mpl1,condVar=TRUE) d2 <- dotplot(r2, par.settings=pp) grid.arrange(d2$gen,d2$popu,nrow=1)
From this plot we can see a hint of regional variability among populations where the Spanish populations (SP) have larger values than Swedish (SW) and Dutch (NL) populations. The difference among genotypes seems to be largely driven by genotype 34.
As seen in Workshop 5, we could have also used the coef() function to plot the random effect predictions or “estimates”, where the sum of random intercept deviation (obtained from ranef()) and the fixed effect (obtained from fixef()) is equal to the parameter estimate obtained from coef().
Random slopes
Final model
We end this section on GLMM with the evaluation of the fixed effects. We first code potential models and compare them using AICc.
- AICc approach
mpl2 <- update(mpl1, . ~ . - rack) # model without rack mpl3 <- update(mpl1, . ~ . - status) # model without status mpl4 <- update(mpl1, . ~ . - amd:nutrient) # without amd:nutrient interaction ICtab(mpl1,mpl2,mpl3,mpl4, type = c("AICc")) # dAICc df # mpl1 0.0 10 # mpl4 1.4 9 # mpl3 1.5 8 # mpl2 55.0 9
Alternatively, we can use the functions drop1 and dfun, where dfun converts the AIC values returned by the drop1 into ΔAIC values (producing a similar table to ICtab above).
- Frequentist approach (drop1)
(dd_LRT <- drop1(mpl1,test="Chisq")) # Model: # total.fruits ~ nutrient * amd + rack + status + (1 | X) + (1 | popu) + (1 | gen) # Df AIC LRT Pr(Chi) # <none> 5015.4 # rack 1 5070.5 57.083 4.179e-14 *** # status 2 5017.0 5.612 0.06044 . # nutrient:amd 1 5016.8 3.444 0.06349 . # --- # Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 (dd_AIC <- dfun(drop1(mpl1))) # Df dAIC # <none> 0.000 # rack 1 55.083 # status 2 1.612 # nutrient:amd 1 1.444
There is a strong effect of rack (a change in AIC of 55 if we remove this variable), whereas the effects of status and of the interaction term are weak (change in AIC when either is dropped is less than 2). We can thus start by removing the non-signifi cant interaction term so that we can test the main e ffects of nutrient and clipping.
- Dropping interaction term
mpl2 <- update(mpl1, . ~ . - and:nutrient) # Use AICc: mpl3 <- update(mpl2, . ~ . - rack) # model without rack or interaction mpl4 <- update(mpl2, . ~ . - status) # model without status or interaction mpl5 <- update(mpl2, . ~ . - nutrient) # without nutrient or interaction mpl6 <- update(mpl2, . ~ . - amd) # without clipping or interaction ICtab(mpl2,mpl3,mpl4,mpl5,mpl6, type = c("AICc")) # dAICc df # mpl2 0.0 9 # mpl4 1.2 7 # mpl6 10.2 8 # mpl3 54.2 8 # mpl5 135.6 8 # Or use drop1: (dd_LRT2 <- drop1(mpl2,test="Chisq")) # Model: # total.fruits ~ nutrient + amd + rack + status + (1 | X) + (1 | popu) + (1 | gen) # Df AIC LRT Pr(Chi) # <none> 5016.8 # nutrient 1 5152.5 137.688 < 2.2e-16 *** # amd 1 5027.0 12.218 0.0004734 *** # rack 1 5071.0 56.231 6.443e-14 *** # status 2 5018.1 5.286 0.0711639 . # --- # Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 (dd_AIC2 <- dfun(drop1(mpl2))) # Df dAIC # <none> 0.000 # nutrient 1 135.688 # amd 1 10.218 # rack 1 54.231 # status 2 1.286 summary(mpl2)
Both the main effects of nutrient and clipping are strong (large change in AIC of 135.6 (mpl5) and 10.2 (mpl6) if either nutrient or clipping are dropped, respectively). Our fi nal model includes the fixed nutrient (strong positive) and clipping (negative) effects, the nuisance variables rack, the observation-level random e ffects (1 | X) to account for over-dispersion and the variation in overall fruit set at the population and genotype levels.
CHALLENGE 7
Using the inverts dataset (larval development times (PLD) of 74 marine invertebrate and vertebrate species reared at different temperatures and time), answer the following questions:
- What is the effect of feeding type and climate (fixed effects) on PLD?
- Does this relationship vary among taxa (random effects)?
- What is the best distribution family for this count data?
- Finally, once you determined the best distribution family, re-evaluate your random and fixed effects.
Additional GLMM resources
Books
B. Bolker (2009) Ecological Models and Data in R. Princeton University Press.
A. Zuur et al. (2009) Mixed Effects Models and Extensions in Ecology with R. Springer.
Websites
GLMM for ecologists: A great website on GLMM with a Q&A section!