QCBS R Workshops
This series of 10 workshops walks participants through the steps required to use R for a wide array of statistical analyses relevant to research in biology and ecology. These open-access workshops were created by members of the QCBS both for members of the QCBS and the larger community.
The content of this workshop has been peer-reviewed by several QCBS members. If you would like to suggest modifications, please contact the current series coordinators, listed on the main wiki page
Workshop 3: Intro to ggplot2, tidyr & dplyr
Developed by: Xavier Giroux-Bougard, Maxwell Farrell, Amanda Winegardner, Étienne Low-Decarie and Monica Granados
Summary: In this workshop we will build on the data manipulation and visualization skills you have learned in base R by introducing three additional R packages: ggplot2, tidyr and dplyr. We’ll learn how to use ggplot2, an excellent plotting alternative to base R that can be used for both diagnostic and publication quality plots. We will then introduce tidyr and dplyr, two powerful tools to manage and re-format your dataset, as well as apply simple or complex functions on subsets of your data. This workshop will be useful for those progressing through the entire workshop series, but also for those who already have some experience in R and would like to become proficient with new tools and packages.
Link to associated Prezi: Prezi
Download the R script for this lesson: Script
1. Plotting in R using the Grammar of Graphics (ggplot2)
Whether you are calculating summary statistics (e.g. Excel), performing more advanced statistical analysis (e.g. JMP, SAS, SPSS), or producing figures and tables (e.g. Sigmaplot, Excel), it is easy to get lost in your workflow when you use a variety of software. This becomes especially problematic every time you import and export your dataset to accomplish a downstream task. With each of these operations, you increase your risk of introducing errors into your data or losing track of the correct version of your data. The R statistical language provides a solution to this by unifying all of the tools you need for advanced data manipulation, statistical analysis, and powerful graphical engines under the same roof. By unifying your workflow from data to tables to figures, you reduce your chances of making mistakes and make your workflow easily understandable and reproducible. Believe us, the future “you” will not regret it! Nor will your collaborators!
1.1 Intro - ggplot2
The most flexible and complete package available for advanced data visualization in R is ggplot2. This packages was created for R by Hadley Wickham based on the Grammar of Graphics by Leland Wilkinson. The source code is hosted on github: https://github.com/hadley/ggplot2. Today we will walk you through the basics of ggplot2, and hopefully its potential will speak for itself and you will have the necessary tools to explore its use for your own projects.
To get started you will need to open RStudio and and load the necessary package from the CRAN repository:
- |
install.packages("ggplot2") require(ggplot2)
1.2 Simple plots using ''qplot()''
Using ''qplot()''
Let's jump right into it then! We will build our first basic plot using the qplot() function in ggplot2. The quick plot function is built to be an intuitive bridge from the plot() function in base R. As such, the syntax is almost identical, and the qplot() function understands how to draw plots based on the types of data (e.g. factor, numerical, etc…) that are mapped. Remember you can always access the help file for a function by typing the command preceded by ?, such as:
- |
?qplot
If you checked the help file we just called, you will see that the 3 first arguments are:
- data
- x
- y
- …
Load data
Before we go forward with qplot(), we need some data to assign values to these arguments. We will first play with the iris dataset, a famous dataset of flower dimensions for three species of iris collected by Edgar Anderson in the Gaspé peninsula right here in Québec! It is already stored as a data.frame directly in R. To load it and explore its structure and the different variables, use the following commands:
Basic scatter plot
Let's build our first scatter plot by mapping the x and y variables from the iris dataset in the qplot() function as follows:
Basic scatter plot (categorical variables)
As mentioned previously, the qplot() function understands how to draw a plot based on the mapped variables. In the previous example we used two numerical variables and obtained a scatter plot. However, qplot() will also understand categorical variables:
Adding axis labels and titles
If you return to the qplot() function's help file, there are several more arguments that can be used to modify different aspects of your figure. Lets start with adding labels and titles using the xlab, ylab and main arguments:
- |
qplot(data = iris, x = Sepal.Length, xlab = "Sepal Length (mm)", y = Sepal.Width, ylab = "Sepal Width (mm)", main = "Sepal dimensions")
ggplot2 - Challenge #1
Using the qplot() function, build a basic scatter plot with a title and axis labels from one of the CO2 or BOD data sets in R. You can load these and explore their contents as follows:
1.3 The Grammar of Graphics
The grammar of graphics is a framework for data visualization that dissects every component of a graph into individual components. Its “awesome factor” is due to this ability, which allows you to flexibly change and modify every single element of a graph. Let's go over a few of these elements, or “layers”, together. First and foremost, a graph requires data, which can be displayed using:
- aesthetics (aes)
- geometric objects (geoms)
- transformations
- axis (coordinate system)
- scales
Aesthetics: ''aes()''
In ggplot2, aesthetics are a group of parameters that specify what and how data is displayed. Here are a few arguments that can be used within the aes() function:
x: position of data along the x axisy: position of data along the y axiscolour: colour of an elementgroup: group that an element belongs toshape: shape used to display a pointlinetype: type of line used (e.g. solid, dashed, etc…)size: size of a point or linealpha: transparency of an element
Geometric objects: ''geoms''
Geometric objects, or geoms, determine the visual representation of your data:
geom_point(): scatterplotgeom_line(): lines connected to points by increasing value of xgeom_path(): lines connected to points in sequence of appearancegeom_boxplot(): box and whiskers plot for categorical variablesgeom_bar(): bar charts for categorical x axisgeom_histogram(): histogram,geom_barfor continuous x axis
How it works
- Create a simple plot object:
plot.object ← ggplot() OR qplot() - Add graphical layers/complexity:
plot.object ← plot.object + layer() - Repeat step 2 until statisfied, then print:
print(plot.object)
Using these steps, we build a final product by laying individual elements over each other until we are satisfied:
Additional resources
Our brief intro on the grammar of graphics is not enough to cover even a small fraction of the all the layers and elements that can be used in visualization. Instead, we introduce the most commonly used, in the hopes that your travels will bring you to the following resources when you take your next steps:
- ''ggplot2'' documentation: this is the best resource for a complete list of available elements and the list of arguments needed for each, as well as useful examples
- SAPE: the Software And Programmer Efficiency research group devotes a full section of their website on appropriate usage of different elements in ggplot2, along with pertinent examples
- The Grammar of Graphics: this book by Leland Wilkinson explains the data visualization framework which ggplot2 uses.
- ggplot2: this book by Hadley Wickham, who authored ggplot2 to implement the grammar of graphics in R.
1.4 Advanced plots using ''ggplot()''
''qplot()'' vs ''ggplot()''
Often, we only require some quick and dirty plots to visualize data, making the qplot() function perfect. As stated above, if we assign a qplot to an object in R, we can still add more complex layers to our base plot like this: plot.object ← plot.object + layer(). However, the qplot() function simply wraps the raw ggplot() commands into a form similar to the plot() function in base R. Let's dissect what ggplot2 is actually doing when we use qplot().
qplot():
- |
qplot(data = iris, x = Sepal.Length, xlab = "Sepal Length (mm)", y = Sepal.Width, ylab = "Sepal Width (mm)", main = "Sepal dimensions")
ggplot():
- |
ggplot(data=iris, aes(x=Sepal.Length, y=Sepal.Width)) + geom_point() + xlab("Sepal Length (mm)") + ylab("Sepal Width (mm)") + ggtitle("Sepal dimensions")
In the raw ggplot() function, we first specify the data and then map the x and y variables in aes(). We subsequently add each individual element one at a time, which unleashes the full potential of grammar of graphics. As your needs shift towards more advanced features in the ggplot2 package, it is good practice to use raw syntax of the ggplot() function, as we will do in the rest of the workshop.
Assign plot to an object
Before we get started with some more advanced features, let's build a foundation by assigning the previous plot to an object:
1.5 Adding colours and shapes
Let's add colours and shapes to our basic scatter plot by adding these as arguments in the aes() function:
It's alive!![]()
1.6 Adding geometric objects
We already have some geometric objects in our basic plot which we added as points using geom_point(). Now we let's add some more advanced geoms, such as linear regressions with geom_smooth():
BONUS
You can even use emojis as your geoms!!! You need the emojiGG package by David Lawrence Miller.
- |
devtools::install_github("dill/emoGG") library(emoGG) #you have to look up the code for the emoji you want emoji_search("bear") 830 bear 1f43b animal 831 bear 1f43b nature 832 bear 1f43b wild ggplot(iris, aes(Sepal.Length, Sepal.Width, color = Species)) + geom_emoji(emoji="1f337")
ggplot2 - Challenge # 2
Produce a colourful plot with linear regression from built in data such as the CO2 dataset or the msleep dataset:
1.7 Adding multiple facets
Data becomes difficult to visualize when there are multiple factors accounted for in experiments. For example, the CO2 data set contains data on CO2 uptake for chilled vs non-chilled treatments in a species of grass from two different regions (Québec and Mississippi). Let's build a basic plot using this data set:
- |
data(CO2) CO2.plot <- ggplot(data = CO2, aes(x=conc, y=uptake, colour= Treatment)) + geom_point() + xlab("CO2 Concentration (mL/L)") + ylab("CO2 Uptake (umol/m^2 sec)") + ggtitle("CO2 uptake in grass plants") print(CO2.plot)
If we want to compare regions, it is useful to make two panels, where the axes are perfectly aligned for data to be compared easily with the human eye. In ggplot2, we can accomplish this with the facet_grid() function. Briefly its basic syntax is as follows: plot.object + facet_grid(rows ~ columns). Here the rows and columns are variables in the data.frame which are factors we want to compare by separating the data into panels. To compare data between regions, we can use the following code:
*Note: you can stack the two panels by using facet.grid(Type ~ .).
1.8 Adding groups
Now that we have two facets, let's observe how the CO2 uptake evolves as CO2 concentrations rise, by adding connecting lines to the points using geom_line():
As we can see from the previous figure, the lines are connecting vertically across three points for each treatment. If we look into the details of the CO2 dataset, this is each treatment in each region has 3 replicates. If we want to plot lines connecting data points for each replicate separately, we can add map a group aesthetic to the “geom_line()” function as follows:
ggplot2 - Challenge # 3
Explore a new geom and other plot elements with your own data or built in data. Look here or here for some inspiration and great examples!
- |
data(msleep) data(OrchardSprays)
1.9 Saving plots
There are many options available to save your beloved creations to file.
Saving plots in RStudio
In RStudio, there are many options available to you to save your figures. You can copy them to the clipboard or export them as any file type (png, jpg, emf, tiff, pdf, metafile, etc…):
Saving plots with code
In instances where you are producing many plots (e.g. during long programs that produces many plots automatically while performing analysis), it is useful to save many plots in one file as a pdf. This can be accomplished as follows:
Saving plots - other options
There are many other options, of particular note is ggsave as it will write directly to your working directory all in one line of code and you can specify the name of the file and the dimensions of the plot:
1.10 Fine tuning - colours
The ggplot2 package automatically chooses colours for you based on the chromatic circle and the number of colours you need. However, in many instances it is useful to use your own colours. You can easily accomplish this using the scale_colour_manual() function:
For added control, you can also use all hexadecimal codes for colours specifically tailored to your needs. A great resource for hex codes is the internets. Just Google “RGB to Hex” and a colour slider will be returned. You can input the hex codes directly into the scale_colour_manual() function instead of colour names.
If you are looking for a colour palette to use across your variables and don't have specific colours in mind. The viridis function will use colour blind and printer friendly colours based on four palettes. You can specify which palette to use, how many colours to use and where to start and end on the spectrum of the palette. Let try adding the default palette “D” to our CO2 plot for the two colours we need.
- |
CO2.plot+scale_colour_manual(values = viridis(2, option = "D"))
BONUS
Check out the following links/packages for some really awesome colour fun:
- Cookbook for R: this ggplot2 and colours entry in the Cookbook for R has tons of great ideas and many color charts and ramps to suggest
- RColorBrewer: this R package is available on the CRAN repository, and is loaded with custom colour ramps which can be integrated to ggplot2 using the
scale_color_brewer()function. - WesAnderson: this r package is available from the github repository, and is loaded with fun and rich color ramps based on themes from your favorite Wes Anderson movies!
1.11 Fine tuning axes and scales
You can control all the aspects of the axes and scales used to display your data (eg break, labels, positions, etc…):
1.12 Fine tuning themes
This is were we get to the “publication quality” part of the ggplot2 package. I am sure that by now, some of you have formed opinions about that grey background… You either love it or hate it (like liver and brussel sprouts). Worry no longer, you will not have to include figures with a grey background in that next publication your are submitting to Nature!
Like every other part of the grammar of graphics, we can modify the theme of the plot to suit our needs, or higher sense of style! Its as simple as:
- |
theme_set(theme())
OR more commonly…
- |
plot.object + theme()
There are way too many theme elements built into the ggplot2 package to mention here, but you can find a complete list in the ggplot theme vignette. Instead of modifying the many elements contained in theme(), you can start from theme functions, which contain a specific set of elements from which to start. For example we can use the “black and white” theme like this:
- |
CO2.plot + theme_bw()
Build your own theme
Another great strategy is to build a theme tailored to your own publication needs, and then apply it to all you figures:
The ggtheme package
The ggtheme package is a great project developed by Jeffrey Arnold on github and also hosted on the CRAN repository, so it can easily be installed as follows:
- |
install.packages('ggthemes', dependencies = TRUE) require(ggthemes)
The package contains many themes, geoms, and colour ramps for ggplot2 which are based on the works of some of the most renown and influential names in the world of data visualization, from the classics such asEdward Tufte to the modern data journalists/programmers at FiveThirtyEight blog.
Here is a quick example which uses the Tufte's boxplots and theme, as you can see he is a minimalist:
- |
data(OrchardSprays) tufte.box.plot <- ggplot(data = OrchardSprays, aes(x = treatment, y = decrease)) + geom_tufteboxplot() + theme_tufte() print(tufte.box.plot)
1.13 ggplot GUI
While hardcore programmers might laugh at you for using a GUI, there is no shame in using them! Jeroen Schouten, who is about as hardcore a programmer as you can get, understood the learning curve for begginners could be steep and so designed an online ggplot2 GUI. While it will not be as fully functional as coding the grammar of graphics, it is very complete. You can import from excel, google spreadsheets, or any data format, and build a few plots using some tutorial videos. The great part is that it shows you the code you have generated to build your figure, which you can copy paste into R as a skeleton on which to add some meat using more advanced features such as themes.

2. Using tidyr to manipulate data frames
2.1 Why "tidy" your data?
Tidying allows you to manipulate the structure of your data while preserving all original information. Many functions in R require or work better with a data structure that isn't the best for readability by people.
In contrast to aggregation, which reduces many cells in the original data set to one cell in the new dataset, tidying preserves a one-to-one connection. Although aggregation can be done with many functions in R, the tiydr package allows you to both reshape and aggregate within a single syntax.
Install / Load the tidyr() package:
- |
if(!require(tidyr)){install.packages("tidyr")} require(tidyr)
Data
In addition to iris and CO2, we will use the built-in datasets “airquality” and “ChickWeight” for this part of the workshop
Explore the datasets:
- |
?airquality str(airquality) head(airquality) names(airquality) ?ChickWeight head(ChickWeight) str(ChickWeight) names(ChickWeight)
You can also use the following code to find other datasets available in R: data()
2.2 Wide vs long data
Let's pretend you send out your field assistant to measure the diameter at breast height (DBH) and height of three tree species for you. The result is this messy (“wide”) data set.
- |
messy <- data.frame( Species = c("Oak", "Elm", "Ash"), DBH = c(12, 20, 13), Height = c(56, 85, 55) View(messy)
“Long” format data has a column for possible variable types and a column for the values of those variables.
The format of your data depends on your specific needs, but some functions and packages such as ggplot2 work well with long format data.
Additionally, long form data can more easily be aggregated and converted back into wide form data to provide summaries, or check the balance of sampling designs.
We can use the tidyr package to:
- 1.“gather” our data (wide –> long)
- 2.“spread” our data (long –> wide)
2.3 Gather: Making your data long
- |
?gather
Most of the packages in the Hadleyverse will require long format data where each row is an entry and each column is a variable. Let's try to “gather” the this messy data using the gather function in tidyr. gather() takes multiple columns, and gathers them into key-value pairs. Note that you have to specify (data, what you want to gather across, the “unit” of your new column, the row identity).
- |
messy.long <- gather(messy, Measurement, cm, -Species) > head(messy.long) Species Measurement cm 1 Oak DBH 12 2 Elm DBH 20 3 Ash DBH 13 4 Oak Height 56 5 Elm Height 85 6 Ash Height 55
Let's try this with the C02 dataset. Here we might want to collapse the last two quantitative variables:
2.4 spread: Making your data wide
Sometimes you might want to go to from long to wide
SPREAD BASICS:spread uses the same syntax as gather (they are complements)
tidyr CHALLENGE
Gather and spread the airquality to return the same format as the original data using month and day.
2.5 separate: Separate two (or more) variables in a single column
Some times you might have really messy data which has two variables in one column. Thankfully the separate function can (wait for it) separate the two variables into two columns
Let's say you have this really messy data set
- |
set.seed(8) really.messy <- data.frame( id = 1:4, trt = sample(rep(c('control', 'farm'), each = 2)), zooplankton.T1 = runif(4), fish.T1 = runif(4), zooplankton.T2 = runif(4), fish.T2 = runif(4) )
First we want to convert this wide dataset to long
- |
really.messy.long <- gather(really.messy, taxa, count, -id, -trt)
Then we want to split those two sampling time (T1 & T2). The syntax we use here is to tell R seperate(data, what column, into what, by what) the tricky part here is telling R where to separate the character string in your column entry using a regular expression to describe the character that separates them.Here the string should be separated by the period (.)
2.6 Combining ggplot with tidyr
Example with the air quality dataset on using both wide and long data formats
- |
head(airquality) Ozone Solar.R Wind Temp Month Day 1 41 190 7.4 67 5 1 2 36 118 8.0 72 5 2 3 12 149 12.6 74 5 3 4 18 313 11.5 62 5 4 5 NA NA 14.3 56 5 5 6 28 NA 14.9 66 5 6
The dataset is in wide format, where measured variables (ozone, solar.r, wind and temp) are placed in their own columns.
Diagnostic plots using the wide format + ggplot2
1: Visualize each individual variable and the range it displays for each month in the timeseries
- |
fMonth <- factor(airquality$Month) #Convert the Month variable to a factor. ozone.box <- ggplot(airquality, aes(x=fMonth, y=Ozone)) + geom_boxplot() solar.box <- ggplot(airquality, aes(x=fMonth, y=Solar.R)) + geom_boxplot() temp.box <- ggplot(airquality, aes(x=fMonth, y=Temp)) + geom_boxplot() wind.box <- ggplot(airquality, aes(x=fMonth, y=Wind)) + geom_boxplot()
You can use grid.arrange() in the package gridExtra to put these plots into 1 figure.
- |
combo.box <- grid.arrange(ozone.box, solar.box, temp.box, wind.box, nrow=2) # nrow = number of rows you would like the plots displayed on.
This arranges the 4 separate plots into one panel for viewing. Note that the scales on the individual y-axes are not the same:
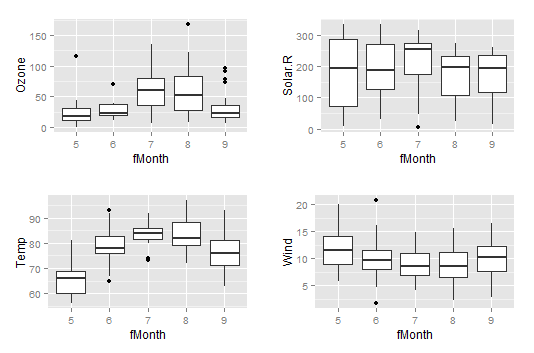
2. You can continue using the wide format of the airquality dataset to make individual plots of each variable showing day measurements for each month.
- |
ozone.plot <- ggplot(airquality, aes(x=Day, y=Ozone)) + geom_point() + geom_smooth() ozone.plot <- ozone.plot + facet_wrap(~Month, nrow=2) solar.plot <- ggplot(airquality, aes(x=Day, y=Solar.R)) + geom_point() +geom_smooth() solar.plot <- solar.plot + facet_wrap(~Month, nrow=2) wind.plot <- ggplot(airquality, aes(x=Day, y=Wind)) + geom_point() +geom_smooth() wind.plot <- wind.plot + facet_wrap(~Month, nrow=2) temp.plot <- ggplot(airquality, aes(x=Day, y=Temp)) + geom_point() +geom_smooth() temp.plot <- temp.plot + facet_wrap(~Month, nrow=2)
You could even then combine these different faceted plots together(though it looks pretty ugly at the moment):

BUT, what if I'd like to use facet_wrap() for the variables as opposed to by month or put all variables on oneplot?
Change data from wide to long format (See back to Section 2.3)
- |
air.long <- gather(airquality, variable, value, -Month, -Day) air.wide <- spread(air.long , variable, value)
Use air.long:
- |
fMonth.long <- factor(air.long$Month) weather <- ggplot(air.long, aes(x=fMonth.long, y=value)) + geom_boxplot() weather <- weather + facet_wrap(~variable, nrow=2)
Compare the weather plot with combo.box

This is the same data but working with it in wide versus long format has allowed us to make different looking plots.
The weather plot uses facet_wrap to put all the individual variables on the same scale. This may be useful in many circumstances. However, using the facet_wrap means that we don't see all the variation present in the wind variable.
In that case, you can modify the code to allow the scales to be determined per facet by setting scales=“free”
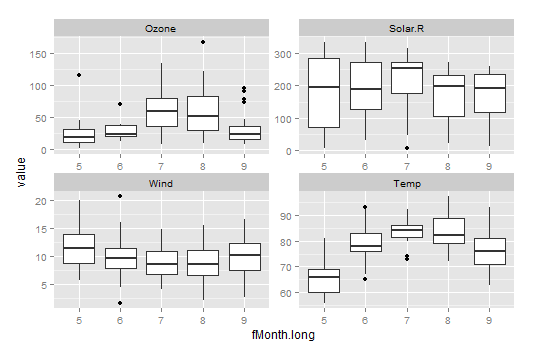
We can also use the long format data (air.long) to create a plot with all the variables included on a single plot:
- |
weather2 <- ggplot(air.long, aes(x=Day, y=value, colour=variable))+ geom_point() #this plot will put all the day measurements on one plot weather2 <- weather2 + facet_wrap(~Month, nrow=1) #add this part and again, the observations are split by month

3. Data manipulation with dplyr
3.1 Intro - the dplyr mission
The vision of the dplyr package is to simplify data manipulation by distilling all the common data manipulation tasks to a set of intuitive verbs. The result is a comprehensive set of tools that allows users to easily translate their thoughts into R code. In addition to ease of use, it is also an amazing package because:
- it can crunch huge datasets wicked fast (written in
Cpp) - it plays nice with the RStudio IDE and other packages in the Hadleyverse
- it can interface with external databases and translate your R code into SQL queries
- if Batman was an R package, he would be
dplyr
3.2 Basic dplyr functions
The dplyr package is built around a core set of “verbs” (or commands). We will start with the following 4 verbs because these operations are ubiquitous in data manipulation:
select(): select columns from a data framefilter(): filter rows according to defined criteriaarrange(): re-order data based on criteria (e.g. ascending, descending)mutate(): create or transform values in a column
Let's load the dplyr package and explore these functions:
- |
if(!require(dplyr)){install.packages("dplyr")} require(dplyr)
In these examples, we will use the airquality dataset.
Select a subset of columns with ''select()''
The airquality dataset contains several columns:
- |
> head(airquality) Ozone Solar.R Wind Temp Month Day 1 41 190 7.4 67 5 1 2 36 118 8.0 72 5 2 3 12 149 12.6 74 5 3 4 18 313 11.5 62 5 4 5 NA NA 14.3 56 5 5 6 28 NA 14.9 66 5 6
Suppose we are only interested in the variation of “Ozone” over time, then we can select the subset of required columns for further analysis:
- |
> ozone <- select(airquality, Ozone, Month, Day) > head(ozone) Ozone Month Day 1 41 5 1 2 36 5 2 3 12 5 3 4 18 5 4 5 NA 5 5 6 28 5 6
As you can see the general format for this function is select(dataframe, column1, column2, …). Most dplyr functions will follow a similarly simple syntax.
Select a subset of rows with ''filter()''
A common operation in data manipulation is the extraction of a subset based on specific conditions. For example, in the airquality dataset, suppose we are interested in analyses that focus on the month of August during high temperature events:
- |
> august <- filter(airquality, Month == 8, Temp >= 90) > head(august) Ozone Solar.R Wind Temp Month Day 1 89 229 10.3 90 8 8 2 110 207 8.0 90 8 9 3 NA 222 8.6 92 8 10 4 76 203 9.7 97 8 28 5 118 225 2.3 94 8 29 6 84 237 6.3 96 8 30
The syntax we employed here is filter(dataframe, logical statement 1, logical statement 2, …). Remember that logical statements provide a TRUE or FALSE answer. The filter() function retains all the data for which the statement is TRUE. This can also be applied on characters and factors.
Sort columns with ''arrange()''
In data manipulation, we sometimes need to sort our data (e.g. numerically or alphabetically) for subsequent operations. A common example of this is a time series. First let's use the following code to create a scrambled version of the airquality dataset:
- |
> air_scrambled <- sample_frac(airquality, 1) > head(air_scrambled) Ozone Solar.R Wind Temp Month Day 21 1 8 9.7 59 5 21 42 NA 259 10.9 93 6 11 151 14 191 14.3 75 9 28 108 22 71 10.3 77 8 16 8 19 99 13.8 59 5 8 104 44 192 11.5 86 8 12
Now let's arrange the data frame back into chronological order, sorting by Month then Day:
- |
> air_chron <- arrange(air_scrambled, Month, Day) > head(air_chron) Ozone Solar.R Wind Temp Month Day 1 41 190 7.4 67 5 1 2 36 118 8.0 72 5 2 3 12 149 12.6 74 5 3 4 18 313 11.5 62 5 4 5 NA NA 14.3 56 5 5 6 28 NA 14.9 66 5 6
Note that we can also sort in descending order by placing the target column in desc() inside the arrange() function.
Create and populate columns with ''mutate()''
Besides subsetting or sorting your data frame, you will often require tools to transform your existing data or generate some additional data based on existing variables. For example, suppose we would like to convert the temperature variable form degrees Fahrenheit to degrees Celsius:
- |
> airquality_C <- mutate(airquality, Temp_C = (Temp-32)*(5/9)) > head(airquality_C) Ozone Solar.R Wind Temp Month Day Temp_C 1 41 190 7.4 67 5 1 19.44444 2 36 118 8.0 72 5 2 22.22222 3 12 149 12.6 74 5 3 23.33333 4 18 313 11.5 62 5 4 16.66667 5 NA NA 14.3 56 5 5 13.33333 6 28 NA 14.9 66 5 6 18.88889
Note that the syntax here is quite simple, but within a single call of the mutate() function, we can replace existing columns, we can create multiple new columns, and each new column can be created using newly created columns within the same function call.
3.3 dplyr and magrittr, a match made in heaven
The magrittr package brings a new and exciting tool to the table: a pipe operator. Pipe operators provide ways of linking functions together so that the output of a function flows into the input of next function in the chain. The syntax for the magrittr pipe operator is %>%. The magrittr pipe operator truly unleashes the full power and potential of dplyr, and we will be using it for the remainder of the workshop. First, let's install and load it:
- |
if(!require(magrittr)){install.packages("magrittr")} require(magrittr)
Using it is quite simple, and we will demonstrate that by combining some of the examples used above. Suppose we wanted to filter() rows to limit our analysis to the month of June, then convert the temperature variable to degrees Celsius. We can tackle this problem step by step, as before:
- |
june_C <- mutate(filter(airquality, Month == 6), Temp_C = (Temp-32)*(5/9))
This code can be difficult to decipher because we start on the inside and work our way out. As we add more operations, the resulting code becomes increasingly illegible. Instead of wrapping each function one inside the other, we can accomplish these 2 operations by linking both functions together:
- |
june_C <- airquality %>% filter(Month == 6) %>% mutate(Temp_C = (Temp-32)*(5/9))
Notice that within each function, we have removed the first argument which specifies the dataset. Instead, we specify our dataset first, then “pipe” into the next function in the chain. This is similar to ggplot2, in that we only specify the data frame once, not every single time we are adding a layer. The advantages of this approach are that our code is less redundant and functions are executed in the same order we read and write them, which makes its easier and quicker to both translate our thoughts into code and read someone else's code and grasp what is being accomplished. As the complexity of your data manipulations increases, it becomes quickly apparent why this is a powerful and elegant approach to writing your dplyr code.
Quick tip: In RStudio we can insert this pipe quickly using the following hotkey: Ctrl (or Cmd for Mac) +Shift+M.
3.4 dplyr - Summaries and grouped operations
The dplyr verbs we have explored so far can be useful on their own, but they become especially powerful when we link them with each other using the pipe operator (%>%) and by applying them to groups of observations. The following functions allow us to split our data frame into distinct groups on which we can then perform operations individually, such as aggregating/summarising:
group_by(): group data frame by a factor for downstream commands (usually summarise)summarise(): summarise values in a data frame or in groups within the data frame with aggregation functions (e.g.min(),max(),mean(), etc…)
These verbs provide the needed backbone for the Split-Apply-Combine strategy that was initially implemented in the plyr package on which dplyr is built. Let's demonstrate the use of these with an example using the airquality dataset. Suppose we are interested in the mean temperature and standard deviation within each month:
- |
> month_sum <- airquality %>% group_by(Month) %>% summarise(mean_temp = mean(Temp), sd_temp = sd(Temp)) > month_sum Source: local data frame [5 x 3] Month mean_temp sd_temp (int) (dbl) (dbl) 1 5 65.54839 6.854870 2 6 79.10000 6.598589 3 7 83.90323 4.315513 4 8 83.96774 6.585256 5 9 76.90000 8.355671
dplyr CHALLENGE
Using the ChickWeight dataset, create a summary table which displays the weight gain of individual chicks in the study. Employ dplyr verbs and the %>% operator.
Ninja Hint
Note that we can group the data frame using more than one factor, using the general syntax as follows: group_by(group1, group2, …). This approach allows us to carry out operations on group2 while conserving information on group1 within the summary table.
dplyr NINJA CHALLENGE
Using the ChickWeight dataset, create a summary table which displays the average weight gain of individual chicks for each diet in the study. Employ dplyr verbs and the %>% operator.
Given that the solution to the last challenge requires that we compute several operations in sequence, it provides a nice example to demonstrate why the syntax implemented by dplyr and magrittr. An additional challenge if you are well versed in base R functions would to reproduce the same operations using fewer key strokes. We tried, and failed… Perhaps we are too accustomed to dplyr now.
3.5 dplyr - Merging data frames
In addition to all the operations we have explored, dplyr also provides some functions that allow you to join two data frames together. The syntax in these functions is simple relative to alternatives in other R packages:
left_join()right_join()inner_join()anti_join()
These are beyond the scope of the current introductory workshop, but they provide extremely useful functionality you may eventually require for some more advanced data manipulation needs.
4. Resources
Here are some great resources for learning ggplot2, tidyr and dplyr that we used when compiling this workshop:
ggplot2
Lecture notes from Hadley's course Stat 405 (the other lessons are awesome too!)
dplyr and tidyr
BONUS! Check out R style guides to help format your scripts for easy reading:
























